Hepatitis B virus X protein interferes with cellular DNA repair
- PMID: 9420223
- PMCID: PMC109372
- DOI: 10.1128/JVI.72.1.266-272.1998
Hepatitis B virus X protein interferes with cellular DNA repair
Abstract
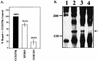
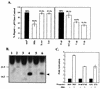
The hepatitis B virus X protein (HBx) is a broadly acting transactivator implicated in the development of liver cancer. Recently, HBx has been reported to interact with several different cellular proteins, including our report of its binding to XAP-1, the human homolog of the simian repair protein UVDDB. In the present study, several HBx mutants were used to localize the minimal domain of HBx required for binding to XAP-1/UVDDB to amino acids 55 to 101. The normal function of XAP-1/UVDDB is thought to involve binding to damaged DNA, the first step in nucleotide excision repair (NER); therefore, we hypothesized that this interaction may affect the cell's capacity to correct lesions in the genome. When tested in two independent assays that measure NER (unscheduled DNA synthesis and host cell reactivation), the expression of HBx significantly inhibited the ability of cells to repair damaged DNA. Under the assay conditions, HBx was expressed at a level similar to that previously observed during natural viral infection and was able to transactivate several target reporter genes. These results are consistent with a model in which HBx acts as a cofactor in hepatocarcinogenesis by preventing the cell from efficiently repairing damaged DNA, thus leading to an accumulation of DNA mutations and, eventually, cancer. An adverse effect on cellular DNA repair processes suggests a new mechanism by which a tumor-associated virus might contribute to carcinogenesis.
Figures


References
-
- Aboussekhra A, Biggerstaff M, Shivji M K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
-
- Aden D P, Fogel A, Plotkin S, Damjanov I, Knowles B B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature (London) 1979;282:615–616. - PubMed
-
- Anonymous. Statistical Package for the Social Sciences (SPSS) for Windows, version 7. Chicago, Ill: SPSS, Inc.; 1995.
-
- Arii M, Takada S, Koike K. Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene. 1992;7:397–403. - PubMed
-
- Athas W F, Hedayati M A, Matanoski G M, Farmer E R, Grossman L. Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res. 1991;51:5786–5793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

