Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2
- PMID: 9420229
- PMCID: PMC109378
- DOI: 10.1128/JVI.72.1.309-319.1998
Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2
Abstract
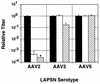
Adeno-associated viruses (AAVs) are single-stranded dependent parvoviruses being developed as transducing vectors. Although at least five serotypes exist (AAV types 1 to 5 [AAV1 to -5]), only AAV2, AAV3, and AAV4 have been sequenced, and the vectors in use were almost all derived from AAV2. Here we report the cloning and sequencing of a second AAV3 genome and a new AAV serotype designated AAV6 that is related to AAV1. AAV2, AAV3, and AAV6 were 82% identical at the nucleotide sequence level, and AAV4 was 75 to 78% identical to these AAVs. Significant sequence variation was noted in portions of the capsid proteins that presumably are responsible for serotype-specific functions. Vectors produced from AAV3 and AAV6 differed from AAV2 vectors in host range and serologic reactivity. The AAV3 and AAV6 vector serotypes were able to transduce cells in the presence of serum from animals previously exposed to AAV2 vectors. Our results suggest that vectors based on alternative AAV serotypes will have advantages over existing AAV2 vectors, including the transduction of different cell types, and resistance to neutralizing antibodies against AAV2. This could be especially important for gene therapy, as significant immunity against AAV2 exists in human populations and many protocols will likely require multiple vector doses.
Figures
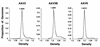






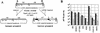
References
-
- Alexander I E, Russell D W, Spence A M, Miller A D. Effects of gamma irradiation on the transduction of dividing and nondividing cells in brain and muscle of rats by adeno-associated virus vectors. Hum Gene Ther. 1996;7:841–850. - PubMed
-
- Atchison R W, Casto B C, Hammon W M. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. - PubMed
-
- Bantel Schaal U, zur Hausen H. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology. 1984;134:52–63. - PubMed
-
- Bertran J, Miller J L, Yang Y, Fenimore-Justman A, Rueda F, Vanin E F, Nienhuis A W. Recombinant adeno-associated virus-mediated high-efficiency, transient expression of the murine cationic amino acid transporter (ecotropic retroviral receptor) permits stable transduction of human HeLa cells by ecotropic retroviral vectors. J Virol. 1996;70:6759–6766. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

