Characterization of the CBF2 binding site within the Epstein-Barr virus latency C promoter and its role in modulating EBNA2-mediated transactivation
- PMID: 9420275
- PMCID: PMC109424
- DOI: 10.1128/JVI.72.1.693-700.1998
Characterization of the CBF2 binding site within the Epstein-Barr virus latency C promoter and its role in modulating EBNA2-mediated transactivation
Abstract
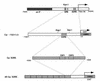
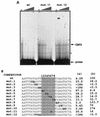
The Epstein-Barr virus (EBV) EBNA2 protein is a transcriptional activator that regulates viral and cellular gene expression and is also essential for EBV-driven immortalization of B lymphocytes. The EBNA2-responsive enhancer in the viral latency C promoter (Cp) binds two cellular factors, CBF1 and CBF2. The precise role of the CBF2 protein for Cp enhancer function is presently unclear. CBF2 does not appear to interact with EBNA2 and binds just downstream of CBF1 between positions -339 and -368 in the Cp EBNA2 enhancer. Within this region an 8-bp sequence, CAGTGCGT, can be found, and a similar sequence is also located downstream of CBF1 binding sites in other EBNA2-responsive promoters. Previous studies have indicated that mutations and methylation in this sequence affect EBNA2 responsiveness. To investigate the requirements for CBF2 binding, we synthesized a series of oligonucleotides carrying double transversion mutations spanning both the conserved core sequence and outside flanking sequences. Surprisingly, mutations outside of the conserved core sequence in 4 bases immediately flanking the 5' end, GGTT, had the most deleterious effect on CBF2 binding. Mutations in the conserved core had a gradient effect, with those near the 5' end having the most deleterious effects on CBF2 binding. In addition, the affinities of CBF2 for binding to the LMP-1, LMP-2, and CD23 promoters were also measured. These promoters contain the conserved core but lack the 5' flanking GGTT motif and bound CBF2 weakly or not at all. Using Cp reporter plasmids containing CBF2 mutant binding sites, we were also able to show that at lower doses of EBNA2, Cp transactivation required a functional CBF2 binding site but that higher doses of EBNA2 transactivated CBF2 mutant promoters to 40% of wild-type levels. These assays indicate that CBF2 is important for EBNA2-mediated transactivation of the viral latency Cp. In addition, CBF2 activity was found to be associated with two polypeptides of 27 and 33 kDa.
Figures
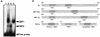
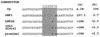

Similar articles
-
Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway.J Virol. 2000 Sep;74(17):8166-75. doi: 10.1128/jvi.74.17.8166-8175.2000. J Virol. 2000. PMID: 10933728 Free PMC article.
-
EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1.J Virol. 1994 Sep;68(9):5375-83. doi: 10.1128/JVI.68.9.5375-5383.1994. J Virol. 1994. PMID: 8057421 Free PMC article.
-
Transcriptional activation signals found in the Epstein-Barr virus (EBV) latency C promoter are conserved in the latency C promoter sequences from baboon and Rhesus monkey EBV-like lymphocryptoviruses (cercopithicine herpesviruses 12 and 15).J Virol. 1999 Jan;73(1):826-33. doi: 10.1128/JVI.73.1.826-833.1999. J Virol. 1999. PMID: 9847397 Free PMC article.
-
Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa.Immunobiology. 1997 Dec;198(1-3):299-306. doi: 10.1016/s0171-2985(97)80050-2. Immunobiology. 1997. PMID: 9442401 Review.
-
EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes.Semin Cancer Biol. 2001 Dec;11(6):423-34. doi: 10.1006/scbi.2001.0409. Semin Cancer Biol. 2001. PMID: 11669604 Review.
Cited by
-
Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway.J Virol. 2000 Sep;74(17):8166-75. doi: 10.1128/jvi.74.17.8166-8175.2000. J Virol. 2000. PMID: 10933728 Free PMC article.
-
Cell cycle association of the retinoblastoma protein Rb and the histone demethylase LSD1 with the Epstein-Barr virus latency promoter Cp.J Virol. 2008 Apr;82(7):3428-37. doi: 10.1128/JVI.01412-07. Epub 2008 Jan 23. J Virol. 2008. PMID: 18216119 Free PMC article.
-
Regulation of Sp100A subnuclear localization and transcriptional function by EBNA-LP and interferon.J Interferon Cytokine Res. 2008 Nov;28(11):667-78. doi: 10.1089/jir.2008.0023. J Interferon Cytokine Res. 2008. PMID: 18844582 Free PMC article.
-
Neoplastic transformation by Notch requires nuclear localization.Mol Cell Biol. 2000 Jun;20(11):3928-41. doi: 10.1128/MCB.20.11.3928-3941.2000. Mol Cell Biol. 2000. PMID: 10805736 Free PMC article.
-
Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter.Mol Cell Biol. 2003 Jun;23(12):4056-65. doi: 10.1128/MCB.23.12.4056-4065.2003. Mol Cell Biol. 2003. PMID: 12773551 Free PMC article.
References
-
- Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. - PubMed
-
- Allday M J, Crawford D H, Griffin B E. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J Gen Virol. 1989;70:1755–1764. - PubMed
-
- Ambinder, R. F. Personal communication.
-
- Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:657–659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

