New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB
- PMID: 9420329
- PMCID: PMC316406
- DOI: 10.1101/gad.12.1.34
New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB
Abstract
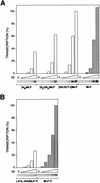
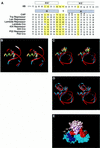
A sequence element located immediately upstream of the TATA element, and having the consensus sequence 5'-G/C-G/C-G/A-C-G-C-C-3', affects the ability of transcription factor IIB to enter transcription complexes and support transcription initiation. The sequence element is recognized directly by the transcription factor IIB. Recognition involves alpha-helices 4' and 5' of IIB, which comprise a helix-turn-helix DNA-binding motif. These observations establish that transcription initiation involves a fourth core promoter element, the IIB recognition element (BRE), in addition to the TATA element, the initiator element, and the downstream promoter element, and involves a second sequence-specific general transcription factor, IIB, in addition to transcription factor IID.
Figures



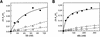
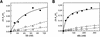

Similar articles
-
The structural basis for the oriented assembly of a TBP/TFB/promoter complex.Proc Natl Acad Sci U S A. 1999 Nov 23;96(24):13668-73. doi: 10.1073/pnas.96.24.13668. Proc Natl Acad Sci U S A. 1999. PMID: 10570130 Free PMC article.
-
Core promoter elements recognized by transcription factor IIB.Biochem Soc Trans. 2006 Dec;34(Pt 6):1051-3. doi: 10.1042/BST0341051. Biochem Soc Trans. 2006. PMID: 17073748
-
Perspectives on the RNA polymerase II core promoter.Biochem Soc Trans. 2006 Dec;34(Pt 6):1047-50. doi: 10.1042/BST0341047. Biochem Soc Trans. 2006. PMID: 17073747
-
X-ray crystallographic studies of eukaryotic transcription initiation factors.Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):483-9. doi: 10.1098/rstb.1996.0046. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735270 Review.
-
The DPE, a core promoter element for transcription by RNA polymerase II.Exp Mol Med. 2002 Sep 30;34(4):259-64. doi: 10.1038/emm.2002.36. Exp Mol Med. 2002. PMID: 12515390 Review.
Cited by
-
ElemeNT: a computational tool for detecting core promoter elements.Transcription. 2015;6(3):41-50. doi: 10.1080/21541264.2015.1067286. Transcription. 2015. PMID: 26226151 Free PMC article.
-
A core promoter element downstream of the TATA box that is recognized by TFIIB.Genes Dev. 2005 Oct 15;19(20):2418-23. doi: 10.1101/gad.342405. Genes Dev. 2005. PMID: 16230532 Free PMC article.
-
Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB.Genes Dev. 2001 Nov 15;15(22):2945-9. doi: 10.1101/gad.206901. Genes Dev. 2001. PMID: 11711430 Free PMC article.
-
Specific down-regulation of an engineered human cyclin D1 promoter by a novel DNA-binding ligand in intact cells.Nucleic Acids Res. 2001 Feb 1;29(3):652-61. doi: 10.1093/nar/29.3.652. Nucleic Acids Res. 2001. PMID: 11160886 Free PMC article.
-
Roles for non-TATA core promoter sequences in transcription and factor binding.Mol Cell Biol. 2000 May;20(10):3608-15. doi: 10.1128/MCB.20.10.3608-3615.2000. Mol Cell Biol. 2000. PMID: 10779350 Free PMC article.
References
-
- Bagby S, Kim S, Maldonado E, Tonk K, Reinberg D, Ikura M. Solution structure of the carboxy-terminal core domain of human TFIIB: Similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. - PubMed
-
- Beamer L, Pabo C. Refined 1.8 Å crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992;227:177–196. - PubMed
-
- Blackwell T, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. - PubMed
-
- Blackwell T, Kretzner L, Blackwood E, Eisenman R, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. - PubMed
-
- Bucher P. The Eukaryotic Promoter Database EPD, EMBL, nucleotide sequence data library, release 48. Cambridge, UK: European Bioinformatics Institute; 1996.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
