The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain
- PMID: 9420330
- PMCID: PMC316402
- DOI: 10.1101/gad.12.1.45
The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain
Abstract
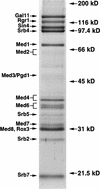
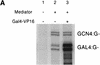
Mediator was resolved from yeast as a multiprotein complex on the basis of its requirement for transcriptional activation in a fully defined system. Three groups of mediator polypeptides could be distinguished: the products of five SRB genes, identified as suppressors of carboxy-terminal domain (CTD)-truncation mutants; products of four genes identified as global repressors; and six members of a new protein family, termed Med, thought to be primarily responsible for transcriptional activation. Notably absent from the purified mediator were Srbs 8, 9, 10, and 11, as well as members of the SWI/SNF complex. The CTD was required for function of mediator in vitro, in keeping with previous indications of involvement of the CTD in transcriptional activation in vivo. Evidence for human homologs of several mediator proteins, including Med7, points to similar mechanisms in higher cells.
Figures







References
-
- Boeke J, Truehart J, Natsoulis B, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
-
- Bröhl S, Lisowsky T, Riemen G, Michaelis G. A new nuclear suppressor system for a mitochondrial RNA polymerase mutant identifies an unusual zinc-finger protein and a polyglutamine domain protein in Saccharomyces cerevisiae. Yeast. 1994;10:719–731. - PubMed
-
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. - PubMed
-
- Chao DM, Gadbois EL, Murray PJ, Anderson SF, Sonu MS, Parvin JD, Young RA. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
