A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm
- PMID: 9420331
- PMCID: PMC316398
- DOI: 10.1101/gad.12.1.55
A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm
Abstract
The SR proteins constitute a large family of nuclear phosphoproteins required for constitutive pre-mRNA splicing. These factors also have global, concentration-dependent effects on alternative splicing regulation and this activity is antagonized by members of the hnRNP A/B family of proteins. We show here that whereas some human SR proteins are confined to the nucleus, three of them-SF2/ASF, SRp20, and 9G8-shuttle rapidly and continuously between the nucleus and the cytoplasm. By swapping the corresponding domains between shuttling and nonshuttling SR proteins, we show that the carboxy-terminal arginine/serine-rich (RS) domain is required for shuttling. This domain, however, is not sufficient to promote shuttling of an unrelated protein reporter, suggesting that stable RNA binding mediated by the RNA-recognition motifs may be required for shuttling. Consistent with such a requirement, a double point-mutation in RRM1 of SF2/ASF that impairs RNA binding prevents the protein from shuttling. In addition, we show that phosphorylation of the RS domain affects the shuttling properties of SR proteins. These findings show that different SR proteins have unique intracellular transport properties and suggest that the family members that shuttle may have roles not only in nuclear pre-mRNA splicing but also in mRNA transport, cytoplasmic events, and/or processes that involve communication between the nucleus and the cytoplasm.
Figures
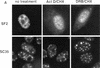

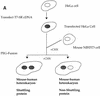




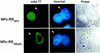
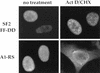

References
-
- Alzhanova-Ericsson AT, Sun X, Visa N, Kiseleva E, Wurtz T, Daneholt B. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes & Dev. 1996;10:2881–2893. - PubMed
-
- Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
