RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli
- PMID: 9422595
- PMCID: PMC106851
- DOI: 10.1128/JB.180.1.73-82.1998
RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli
Abstract
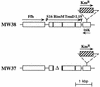
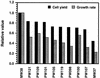
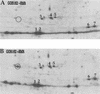
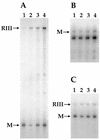
The trmD operon is located at 56.7 min on the genetic map of the Escherichia coli chromosome and contains the genes for ribosomal protein (r-protein) S16, a 21-kDa protein (RimM, formerly called 21K), the tRNA (m1G37)methyltransferase (TrmD), and r-protein L19, in that order. Previously, we have shown that strains from which the rimM gene has been deleted have a sevenfold-reduced growth rate and a reduced translational efficiency. The slow growth and translational deficiency were found to be partly suppressed by mutations in rpsM, which encodes r-protein S13. Further, the RimM protein was shown to have affinity for free ribosomal 30S subunits but not for 30S subunits in the 70S ribosomes. Here we have isolated several new suppressor mutations, most of which seem to be located close to or within the nusA operon at 68.9 min on the chromosome. For at least one of these mutations, increased expression of the ribosome binding factor RbfA is responsible for the suppression of the slow growth and translational deficiency of a deltarimM mutant. Further, the RimM and RbfA proteins were found to be essential for efficient processing of 16S rRNA.
Figures




References
-
- Apirion D, Miczak A. RNA processing in prokaryotic cells. Bioessays. 1993;15:113–120. - PubMed
-
- Björk G R. Modification of stable RNA. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 719–731.
-
- Björk G R, Ericson J U, Gustafsson C E, Hagervall T G, Jönsson Y H, Wikström P M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

