Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells
- PMID: 9423848
- PMCID: PMC107867
- DOI: 10.1128/IAI.66.1.122-131.1998
Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells
Abstract
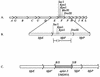
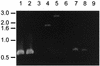
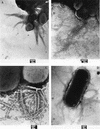
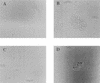
Adherence of enteropathogenic Escherichia coli (EPEC) to epithelial cells is dependent on a type IV fimbria, termed the bundle-forming pilus (BFP). A cluster of 14 genes is required for expression of BFP. The eighth gene in the cluster, bfpF, encodes a putative nucleotide-binding protein which resembles the PilT protein of Pseudomonas aeruginosa. It has been proposed that PilT is required for the retraction of the P. aeruginosa pilus, which results in twitching motility. To test the role of BfpF in BFP function and EPEC pathogenesis, two different mutations were constructed in the bfpF gene, one in the cloned gene cluster in a laboratory E. coli strain and one in wild-type EPEC. Neither mutation affected prepilin synthesis, leader sequence processing, or pilus biogenesis. However, both mutations resulted in increased localized adherence. In addition, the EPEC bfpF mutant displayed increased aggregation. The EPEC bfpF mutant was not deficient in attaching and effacing activity or invasion capacity. These results suggest that BfpF decreases aggregation and adherence by EPEC but that subsequent steps in EPEC pathogenesis do not require this protein.
Figures



References
-
- Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. - PubMed
-
- Alm R A, Mattick J S. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol. 1995;16:485–496. - PubMed
-
- Baldini M M, Kaper J B, Levine M M, Candy D C, Moon H W. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J Pediatr Gastroenterol Nutr. 1983;2:534–538. - PubMed
-
- Bradley D E. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology. 1974;58:149–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

