Repression of phenol catabolism by organic acids in Ralstonia eutropha
- PMID: 9435054
- PMCID: PMC124663
- DOI: 10.1128/AEM.64.1.1-6.1998
Repression of phenol catabolism by organic acids in Ralstonia eutropha
Abstract
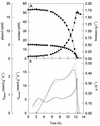
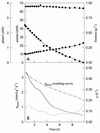
During batch growth of Ralstonia eutropha (previously named Alcaligenes eutrophus) on phenol in the presence of acetate, acetate was found to be the preferred substrate; this organic acid was rapidly metabolized, and the specific rate of phenol consumption was considerably decreased, although phenol consumption was not abolished. This decrease corresponded to a drop in phenol hydroxylase and catechol-2,3-dioxygenase specific activities, and the synthesis of the latter was repressed at the transcriptional level. Studies with a mutant not able to consume acetate indicated that the organic acid itself triggers the repression. Other organic acids were also found to repress phenol degradation. One of these, benzoate, was found to completely block the catabolism of phenol (diauxic growth). A mutant unable to metabolize benzoate was also unable to develop on benzoate-phenol mixtures, indicating that the organic acid rather than a metabolite involved in benzoate degradation was responsible for the repression observed.
Figures



References
-
- Ampe F, Lindley N D. Flux limitations in the ortho pathway of benzoate degradation of Alcaligenes eutrophus: metabolite overflow and induction of the meta pathway at high substrate concentrations. Microbiology. 1996;142:1807–1817. - PubMed
-
- Ampe F, Léonard D, Lindley N D. Growth performance and pathway flux determine substrate preference of Alcaligenes eutrophus during growth on acetate plus aromatic compound mixtures. Appl Microbiol Biotechnol. 1996;46:562–569.
-
- Babel W, Brinkmann U, Mueller R H. The auxiliary substrate concept. An approach for overcoming limits of microbial performances. Acta Biotechnol. 1993;13:211–242.
-
- Bartilson M, Shingler V. Nucleotide sequence and expression of the catechol 2,3-dioxygenase-encoding gene of phenol-catabolizing Pseudomonas CF600. Gene. 1989;85:233–238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

