Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes
- PMID: 9435219
- PMCID: PMC18447
- DOI: 10.1073/pnas.95.2.492
Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes
Abstract
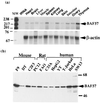
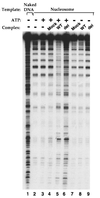
The SWI/SNF complex in yeast and Drosophila is thought to facilitate transcriptional activation of specific genes by antagonizing chromatin-mediated transcriptional repression. The mechanism by which it is targeted to specific genes is poorly understood and may involve direct DNA binding and/or interactions with specific or general transcription factors. We have previously purified a mammalian complex by using antibodies against BRG1, a human homologue of SWI2/SNF2. This complex is likely functionally related to the yeast SWI/SNF complex because all five subunit identified so far (referred to as BAFs, for BRG1-associated factors) are homologues of the yeast SWI/SNF subunits. However, we now describe the cloning of the 57-kDa subunit (BAF57), which is present only in higher eukaryotes but not in yeast. BAF57 is shared by all mammalian complexes and contains a high-mobility-group (HMG) domain adjacent to a kinesin-like region. Both recombinant BAF57 and the whole complex bind four-way junction (4WJ) DNA, which is thought to mimic the topology of DNA as it enters or exits the nucleosome. Surprisingly, complexes with mutations in the HMG domain of BAF57 can still bind 4WJ DNA and mediate ATP-dependent nucleosome disruption. Our work describes the first DNA binding subunit for SWI/SNF-like complexes and suggest that the mechanism by which mammalian and Drosophila SWI/SNF-like complexes interact with chromatin may involve recognition of higher-order chromatin structure by two or more DNA binding domains.
Figures





Similar articles
-
Diversity and specialization of mammalian SWI/SNF complexes.Genes Dev. 1996 Sep 1;10(17):2117-30. doi: 10.1101/gad.10.17.2117. Genes Dev. 1996. PMID: 8804307
-
Functional selectivity of recombinant mammalian SWI/SNF subunits.Genes Dev. 2000 Oct 1;14(19):2441-51. doi: 10.1101/gad.828000. Genes Dev. 2000. PMID: 11018012 Free PMC article.
-
Purification and biochemical heterogeneity of the mammalian SWI-SNF complex.EMBO J. 1996 Oct 1;15(19):5370-82. EMBO J. 1996. PMID: 8895581 Free PMC article.
-
The SWI/SNF complex and cancer.Oncogene. 2009 Apr 9;28(14):1653-68. doi: 10.1038/onc.2009.4. Epub 2009 Feb 23. Oncogene. 2009. PMID: 19234488 Review.
-
The developmental and pathogenic roles of BAF57, a special subunit of the BAF chromatin-remodeling complex.FEBS Lett. 2016 Jun;590(11):1555-69. doi: 10.1002/1873-3468.12201. Epub 2016 May 20. FEBS Lett. 2016. PMID: 27149204 Review.
Cited by
-
ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF.Mol Cell Biol. 2013 Jan;33(2):265-80. doi: 10.1128/MCB.01008-12. Epub 2012 Nov 5. Mol Cell Biol. 2013. PMID: 23129809 Free PMC article.
-
Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner.Mol Cell Biol. 2003 Apr;23(8):2942-52. doi: 10.1128/MCB.23.8.2942-2952.2003. Mol Cell Biol. 2003. PMID: 12665591 Free PMC article.
-
HuR regulates p21 mRNA stabilization by UV light.Mol Cell Biol. 2000 Feb;20(3):760-9. doi: 10.1128/MCB.20.3.760-769.2000. Mol Cell Biol. 2000. PMID: 10629032 Free PMC article.
-
A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells.Nat Commun. 2018 Dec 3;9(1):5139. doi: 10.1038/s41467-018-07528-9. Nat Commun. 2018. PMID: 30510198 Free PMC article.
-
Cannabichromene Induces Neuronal Differentiation in NSC-34 Cells: Insights from Transcriptomic Analysis.Life (Basel). 2023 Mar 9;13(3):742. doi: 10.3390/life13030742. Life (Basel). 2023. PMID: 36983897 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

