Promoter specificity determinants of T7 RNA polymerase
- PMID: 9435223
- PMCID: PMC18451
- DOI: 10.1073/pnas.95.2.515
Promoter specificity determinants of T7 RNA polymerase
Abstract
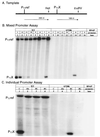
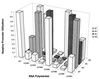
The high specificity of T7 RNA polymerase (RNAP) for its promoter sequence is mediated, in part, by a specificity loop (residues 742-773) that projects into the DNA binding cleft (1). Previous work demonstrated a role for the amino acid residue at position 748 (N748) in this loop in discrimination of the base pairs (bp) at positions -10 and -11 (2). A comparison of the sequences of other phage RNAPs and their promoters suggested additional contacts that might be important in promoter recognition. We have found that changing the amino acid residue at position 758 in T7 RNAP results in an enzyme with altered specificity for the bp at position -8. The identification of two amino acid:base pair contacts (i.e., N748 with the bp at -10 and -11, and Q758 with the bp at -8) provides information concerning the disposition of the specificity loop relative to the upstream region of the promoter. The results suggest that substantial rearrangements of the loop (and/or the DNA) are likely to be required to allow these amino acids to interact with their cognate base pairs during promoter recognition.
Figures


Similar articles
-
Substitution of a single bacteriophage T3 residue in bacteriophage T7 RNA polymerase at position 748 results in a switch in promoter specificity.J Mol Biol. 1992 Nov 20;228(2):506-15. doi: 10.1016/0022-2836(92)90838-b. J Mol Biol. 1992. PMID: 1453460
-
T7 RNA polymerase mutants with altered promoter specificities.Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3147-51. doi: 10.1073/pnas.90.8.3147. Proc Natl Acad Sci U S A. 1993. PMID: 8475053 Free PMC article.
-
Probing the interaction of T7 RNA polymerase with promoter.Biochemistry. 1999 Apr 20;38(16):4972-81. doi: 10.1021/bi9823941. Biochemistry. 1999. PMID: 10213599
-
Structural-functional analysis of bacteriophage T7 RNA polymerase.Biochemistry (Mosc). 2002 Oct;67(10):1124-35. doi: 10.1023/a:1020911223250. Biochemistry (Mosc). 2002. PMID: 12460110 Review.
-
Structure and function of the bacteriophage T7 RNA polymerase (or, the virtues of simplicity).Cell Mol Biol Res. 1993;39(4):385-91. Cell Mol Biol Res. 1993. PMID: 8312975 Review.
Cited by
-
mRNA vaccines: a new era in vaccine development.Oncol Res. 2024 Sep 18;32(10):1543-1564. doi: 10.32604/or.2024.043987. eCollection 2024. Oncol Res. 2024. PMID: 39308511 Free PMC article. Review.
-
Compatibility and Fidelity of Mirror-Image Thymidine in Transcription Events by T7 RNA Polymerase.Mol Ther Nucleic Acids. 2020 Sep 4;21:604-613. doi: 10.1016/j.omtn.2020.06.023. Epub 2020 Jun 27. Mol Ther Nucleic Acids. 2020. PMID: 32721880 Free PMC article.
-
A model for transcription initiation in human mitochondria.Nucleic Acids Res. 2015 Apr 20;43(7):3726-35. doi: 10.1093/nar/gkv235. Epub 2015 Mar 23. Nucleic Acids Res. 2015. PMID: 25800739 Free PMC article.
-
Promoter Length Affects the Initiation of T7 RNA Polymerase In Vitro: New Insights into Promoter/Polymerase Co-evolution.J Mol Evol. 2020 Mar;88(2):179-193. doi: 10.1007/s00239-019-09922-3. Epub 2019 Dec 21. J Mol Evol. 2020. PMID: 31863129
-
Directed evolution of an orthogonal transcription engine for programmable gene expression in eukaryotes.bioRxiv [Preprint]. 2024 Sep 25:2024.09.25.614978. doi: 10.1101/2024.09.25.614978. bioRxiv. 2024. Update in: iScience. 2024 Dec 06;28(1):111541. doi: 10.1016/j.isci.2024.111541. PMID: 39386662 Free PMC article. Updated. Preprint.
References
-
- Sousa R, Chung Y J, Rose J P, Wang B C. Nature (London) 1993;364:593–599. - PubMed
-
- Raskin C A, Diaz G A, Joho K, McAllister W T. J Mol Biol. 1992;228:506–515. - PubMed
-
- McAllister W T. Cell Mol Biol Res. 1993;39:385–391. - PubMed
-
- Klement J F, Moorefield M B, Jorgensen E D, Brown J E, Risman S, McAllister W T. J Mol Biol. 1990;215:21–29. - PubMed
-
- Lee S S, Kang C W. Biochem Int. 1992;26:1–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

