Initiation and velocity of chromosome replication in Escherichia coli B/r and K-12
- PMID: 9440515
- PMCID: PMC106881
- DOI: 10.1128/JB.180.2.265-273.1998
Initiation and velocity of chromosome replication in Escherichia coli B/r and K-12
Abstract
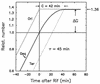
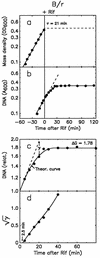
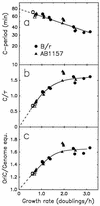
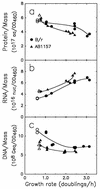
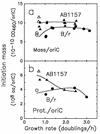
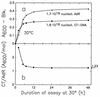
The macromolecular composition and a number of parameters affecting chromosome replication were examined over a range of exponential growth rates in two common Escherichia coli strains, B/r and K-12 AB1157. Based on improved measurements of DNA after treatment of exponential cultures with rifampin, the cell mass per chromosomal replication origin (initiation mass) and the time required to replicate the chromosome from origin to terminus (C period) were determined. For these two strains, the initiation mass approached values of 8 x 10(-10) and 10 x 10(-10) units of optical density (at 460 nm) of culture mass per oriC, respectively, at growth rates above 1 doubling/h (at 37 degrees C). The amount of protein per oriC decreased with increasing growth rate for AB1157 and remained nearly constant for the B/r strain. The C period decreased for both strains in an essentially identical manner from about 70 min at 0.6 doublings/h to about 33 min at 3 doublings/h. From the initiation mass and C period, relative or absolute copy numbers for genes with known map locations can be accurately determined at different growth rates. At growth rates above 2 doublings/h, when chromosomes are highly branched, genes near the origin are about threefold more prevalent than genes near the terminus. At a growth rate of 0.6 doubling/h, this ratio is only about 1.7, which reflects the lower degree of chromosome branching.
Figures


References
-
- Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2460–2488.
-
- Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1715–1902.
-
- Bird R E, Louarn J, Martuscelli J, Caro L. Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol. 1972;70:549–566. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

