Expression cloning of a Pseudomonas gene encoding a hydroxydecanoyl-acyl carrier protein-dependent UDP-GlcNAc acyltransferase
- PMID: 9440522
- PMCID: PMC106888
- DOI: 10.1128/JB.180.2.330-337.1998
Expression cloning of a Pseudomonas gene encoding a hydroxydecanoyl-acyl carrier protein-dependent UDP-GlcNAc acyltransferase
Abstract
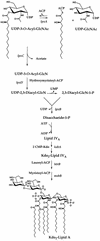
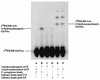
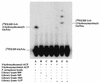
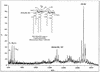
UDP-N-acetylglucosamine-3-O-acyltransferase (UDP-GlcNAc acyltransferase) catalyzes the first step of lipid A biosynthesis (M. S. Anderson and C. R. H. Raetz, J. Biol. Chem. 262:5159-5169, 1987). We here report the isolation of the lpxA gene of Pseudomonas aeruginosa from a library of Pseudomonas strain PAO1 expressed in Escherichia coli LE392 (J. Lightfoot and J. S. Lam, J. Bacteriol. 173:5624-5630, 1991). Pseudomonas lpxA encodes a 10-carbon-specific UDP-GlcNAc acyltransferase, whereas the E. coli transferase is selective for a 14-carbon acyl chain. Recombinant cosmid 1137 enabled production of a 3-hydroxydecanoyl-specific UDP-GlcNAc acyltransferase in E. coli. It was identified by assaying lysozyme-EDTA lysates of individual members of the library with 3-hydroxydecanoyl-acyl carrier protein (ACP) as the substrate. Cosmid 1137 contained a 20-kb insert of P. aeruginosa DNA. The lpxA gene region was localized to a 1.3-kb SalI-PstI fragment. Sequencing revealed that it contains one complete open reading frame (777 bp) encoding a new lpxA homolog. The predicted Pseudomonas LpxA is 258 amino acids long and contains 21 complete hexapeptide repeating units, spaced in approximately the same manner as the 24 repeats of E. coli LpxA. The P. aeruginosa UDP-GlcNAc acyltransferase is 54% identical and 67% similar to the E. coli enzyme. A plasmid (pGD3) containing the 1.3-kb SalI-PstI fragment complemented E. coli RO138, a temperature-sensitive mutant harboring lpxA2. LpxA assays of extracts of this construct indicated that it is > 1,000-fold more selective for 3-hydroxydecanoyl-ACP than for 3-hydroxymyristoyl-ACP. Mass spectrometry of lipid A isolated from this strain by hydrolysis at pH 4.5 revealed [M-H]- 1,684.5 (versus 1,796.5 for wild-type lipid A), consistent with 3-hydroxydecanoate rather than 3-hydroxymyristate at positions 3 and 3'.
Figures



Similar articles
-
Shortened hydroxyacyl chains on lipid A of Escherichia coli cells expressing a foreign UDP-N-acetylglucosamine O-acyltransferase.J Biol Chem. 1997 Aug 8;272(32):19688-96. doi: 10.1074/jbc.272.32.19688. J Biol Chem. 1997. PMID: 9242624
-
Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases.J Biol Chem. 1998 Dec 4;273(49):32369-72. doi: 10.1074/jbc.273.49.32369. J Biol Chem. 1998. PMID: 9829962
-
Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.Biochemistry. 2015 Sep 29;54(38):5937-48. doi: 10.1021/acs.biochem.5b00720. Epub 2015 Sep 18. Biochemistry. 2015. PMID: 26352800
-
Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase.Proc Natl Acad Sci U S A. 2007 Aug 21;104(34):13543-50. doi: 10.1073/pnas.0705833104. Epub 2007 Aug 13. Proc Natl Acad Sci U S A. 2007. PMID: 17698807 Free PMC article.
-
The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis.J Biol Chem. 1993 Sep 15;268(26):19866-74. J Biol Chem. 1993. PMID: 8366125
Cited by
-
The Pseudomonas aeruginosa rhlG gene encodes an NADPH-dependent beta-ketoacyl reductase which is specifically involved in rhamnolipid synthesis.J Bacteriol. 1998 Sep;180(17):4442-51. doi: 10.1128/JB.180.17.4442-4451.1998. J Bacteriol. 1998. PMID: 9721281 Free PMC article.
-
An Eight-Residue Deletion in Escherichia coli FabG Causes Temperature-Sensitive Growth and Lipid Synthesis Plus Resistance to the Calmodulin Inhibitor Trifluoperazine.J Bacteriol. 2017 Apr 25;199(10):e00074-17. doi: 10.1128/JB.00074-17. Print 2017 May 15. J Bacteriol. 2017. PMID: 28264990 Free PMC article.
-
Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function.Microbiol Mol Biol Rev. 1998 Jun;62(2):309-33. doi: 10.1128/MMBR.62.2.309-333.1998. Microbiol Mol Biol Rev. 1998. PMID: 9618444 Free PMC article. Review.
-
Genetic and Functional Diversity of Pseudomonas aeruginosa Lipopolysaccharide.Front Microbiol. 2011 Jun 1;2:118. doi: 10.3389/fmicb.2011.00118. eCollection 2011. Front Microbiol. 2011. PMID: 21687428 Free PMC article.
-
Acyltransferases in bacteria.Microbiol Mol Biol Rev. 2013 Jun;77(2):277-321. doi: 10.1128/MMBR.00010-13. Microbiol Mol Biol Rev. 2013. PMID: 23699259 Free PMC article. Review.
References
-
- Anderson M S, Bulawa C E, Raetz C R H. The biosynthesis of gram-negative endotoxin: formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J Biol Chem. 1985;260:15536–15541. - PubMed
-
- Anderson M S, Bull H S, Galloway S M, Kelly T M, Mohan S, Radika K, Raetz C R H. UDP-N-acetylglucosamine acyltransferase of Escherichia coli: the first step of endotoxin biosynthesis is thermodynamically unfavorable. J Biol Chem. 1993;268:19858–19865. - PubMed
-
- Anderson M S, Raetz C R H. Biosynthesis of lipid A precursors in Escherichia coli: a cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J Biol Chem. 1987;262:5159–5169. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989.
-
- Beaman T W, Binder D A, Blanchard J S, Roderick S L. Three-dimensional structure of tetrahydrodipicolinate N-succinyltransferase. Biochemistry. 1997;36:489–494. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

