Conversion of NfsA, the major Escherichia coli nitroreductase, to a flavin reductase with an activity similar to that of Frp, a flavin reductase in Vibrio harveyi, by a single amino acid substitution
- PMID: 9440535
- PMCID: PMC106901
- DOI: 10.1128/JB.180.2.422-425.1998
Conversion of NfsA, the major Escherichia coli nitroreductase, to a flavin reductase with an activity similar to that of Frp, a flavin reductase in Vibrio harveyi, by a single amino acid substitution
Abstract
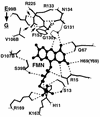
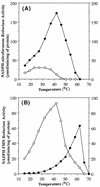
NfsA is the major oxygen-insensitive nitroreductase of Escherichia coli, similar in amino acid sequence to Frp, a flavin reductase of Vibrio harveyi. Here, we show that a single amino acid substitution at position 99, which may destroy three hydrogen bonds in the putative active center, transforms NfsA from a nitroreductase into a flavin reductase that is as active as the authentic Frp and a tartrazine reductase that is 30-fold more active than wild-type NfsA.
Figures

References
-
- Bryant D W, McCalla D R, Leeksma M, Laneuville P. Type I nitroreductases of Escherichia coli. Can J Microbiol. 1981;27:81–86. - PubMed
-
- Duane W, Hastings J W. Flavin mononucleotide reductase of luminous bacteria. Mol Cell Biochem. 1975;6:53–64. - PubMed
-
- Jablonski E, DeLuca M. Purification and properties of the NADH and NADPH specific FMN oxidoreductases from Beneckea harveyi. Biochemistry. 1977;16:2932–2936. - PubMed
-
- Koike, H., H. Sasaki, T. Kobori, S. Zenno, K. Saigo, M. E. P. Murphy, E. T. Adman, and M. Tanokura. Unpublished data. - PubMed
-
- Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

