The pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and retrotransposition in Saccharomyces cerevisiae
- PMID: 9444997
- PMCID: PMC124575
- DOI: 10.1128/JVI.72.2.1036-1042.1998
The pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and retrotransposition in Saccharomyces cerevisiae
Abstract
Programmed ribosomal frameshifting is a molecular mechanism that is used by many RNA viruses to produce Gag-Pol fusion proteins. The efficiency of these frameshift events determines the ratio of viral Gag to Gag-Pol proteins available for viral particle morphogenesis, and changes in ribosomal frameshift efficiencies can severely inhibit virus propagation. Since ribosomal frameshifting occurs during the elongation phase of protein translation, it is reasonable to hypothesize that agents that affect the different steps in this process may also have an impact on programmed ribosomal frameshifting. We examined the molecular mechanisms governing programmed ribosomal frameshifting by using two viruses of the yeast Saccharomyces cerevisiae. Here, we present evidence that pokeweed antiviral protein (PAP), a single-chain ribosomal inhibitory protein that depurinates an adenine residue in the alpha-sarcin loop of 25S rRNA and inhibits translocation, specifically inhibits Ty1-directed +1 ribosomal frameshifting in intact yeast cells and in an in vitro assay system. Using an in vivo assay for Ty1 retrotransposition, we show that PAP specifically inhibits Ty1 retrotransposition, suggesting that Ty1 viral particle morphogenesis is inhibited in infected cells. PAP does not affect programmed -1 ribosomal frameshift efficiencies, nor does it have a noticeable impact on the ability of cells to maintain the M1-dependent killer virus phenotype, suggesting that -1 ribosomal frameshifting does not occur after the peptidyltransferase reaction. These results provide the first evidence that PAP has viral RNA-specific effects in vivo which may be responsible for the mechanism of its antiviral activity.
Figures

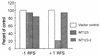

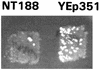
Similar articles
-
Ty1 retrotransposition and programmed +1 ribosomal frameshifting require the integrity of the protein synthetic translocation step.Virology. 2001 Jul 20;286(1):216-24. doi: 10.1006/viro.2001.0997. Virology. 2001. PMID: 11448174
-
A C-terminal deletion mutant of pokeweed antiviral protein inhibits programmed +1 ribosomal frameshifting and Ty1 retrotransposition without depurinating the sarcin/ricin loop of rRNA.Virology. 2001 Jan 5;279(1):292-301. doi: 10.1006/viro.2000.0647. Virology. 2001. PMID: 11145910
-
Frameshift signal transplantation and the unambiguous analysis of mutations in the yeast retrotransposon Ty1 Gag-Pol overlap region.J Virol. 2001 Aug;75(15):6769-75. doi: 10.1128/JVI.75.15.6769-6775.2001. J Virol. 2001. PMID: 11435555 Free PMC article.
-
A review on architecture of the gag-pol ribosomal frameshifting RNA in human immunodeficiency virus: a variability survey of virus genotypes.J Biomol Struct Dyn. 2017 Jun;35(8):1629-1653. doi: 10.1080/07391102.2016.1194231. Epub 2016 Aug 2. J Biomol Struct Dyn. 2017. PMID: 27485859 Review.
-
Ribosomal frameshifting in yeast viruses.Yeast. 1995 Sep 30;11(12):1115-27. doi: 10.1002/yea.320111202. Yeast. 1995. PMID: 8619310 Free PMC article. Review.
Cited by
-
Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae.PLoS One. 2007 Jun 13;2(6):e517. doi: 10.1371/journal.pone.0000517. PLoS One. 2007. PMID: 17565370 Free PMC article.
-
Ribosome depurination is not sufficient for ricin-mediated cell death in Saccharomyces cerevisiae.Infect Immun. 2007 Jan;75(1):417-28. doi: 10.1128/IAI.01295-06. Epub 2006 Nov 13. Infect Immun. 2007. PMID: 17101666 Free PMC article.
-
Structural analysis of the -1 ribosomal frameshift elements in giardiavirus mRNA.J Virol. 2001 Nov;75(22):10612-22. doi: 10.1128/JVI.75.22.10612-10622.2001. J Virol. 2001. PMID: 11602703 Free PMC article.
-
Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2.Nucleic Acids Res. 2001 Mar 1;29(5):1125-31. doi: 10.1093/nar/29.5.1125. Nucleic Acids Res. 2001. PMID: 11222762 Free PMC article.
-
Delayed rRNA processing results in significant ribosome biogenesis and functional defects.Mol Cell Biol. 2003 Mar;23(5):1602-13. doi: 10.1128/MCB.23.5.1602-1613.2003. Mol Cell Biol. 2003. PMID: 12588980 Free PMC article.
References
-
- Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol. 1995;76:1885–1892. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

