Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination
- PMID: 9445017
- PMCID: PMC124595
- DOI: 10.1128/JVI.72.2.1186-1194.1998
Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination
Abstract
Two models for the mechanism of retroviral recombination have been proposed: forced copy choice (minus-strand recombination) and strand displacement-assimilation (plus-strand recombination). Each minus-strand recombination event results in one template switch, whereas each plus-strand recombination event results in two template switches. Recombinant proviruses with one and more than one template switches were previously observed. Recombinants with one template switch were generated by minus-strand recombination, while recombinants containing more than one template switch may have been generated by plus-strand recombination or by correlated minus-strand recombination. We recently observed that retroviral recombination exhibits high negative interference whereby the frequency of recombinants containing multiple template-switching events is higher than expected. To delineate the mechanism that generates recombinants with more than one template switch, we devised a system that permits only minus-strand recombination. Two highly homologous vectors, WH204 and WH221, containing eight different restriction site markers were used. The primer binding site (PBS) of WH221 was deleted; although reverse transcription cannot initiate from WH221 RNA, it can serve as a template for DNA synthesis in heterozygotic virions. After one round of retroviral replication, the structures of the recombinant proviruses were examined. Recombinants containing two, three, four, and five template switches were observed at 1.4-, 10-, 65-, and 50-fold-higher frequencies, respectively, than expected. This indicates that minus-strand recombination events are correlated and can generate proviruses with multiple template switches efficiently. The frequencies of recombinants containing multiple template switches were similar to those observed in the previous system, which allowed both minus- and plus-strand recombination. Thus, the previously reported high negative interference during retroviral recombination can be caused by correlated template switches during minus-strand DNA synthesis. In addition, all examined recombinants contained an intact PBS, indicating that most of the plus-strand DNA transfer occurs after completion of the strong-stop DNA.
Figures


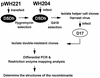
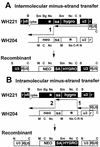

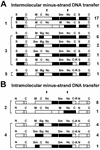
References
-
- Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. - PubMed
-
- Coffin J M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

