Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation
- PMID: 9445018
- PMCID: PMC124596
- DOI: 10.1128/JVI.72.2.1195-1202.1998
Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation
Abstract
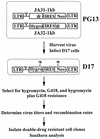
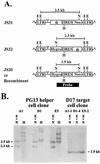
Recombination occurs at high frequencies in all examined retroviruses. The previously determined homologous recombination rate in one retroviral replication cycle is 4% for markers 1.0 kb apart in spleen necrosis virus (SNV). This has often been used to suggest that approximately 30 to 40% of the replication-competent viruses with 7- to 10-kb genomes undergo recombination. These estimates were based on the untested assumption that a linear relationship exists between recombination rates and marker distances. To delineate this relationship, we constructed three sets of murine leukemia virus (MLV)-based vectors containing the neomycin phosphotransferase gene (neo) and the hygromycin phosphotransferase B gene (hygro). Each set contained one vector with a functional neo and an inactivated hygro and one vector with a functional hygro and an inactivated neo. The two inactivating mutations in the three sets of vectors were separated by 1.0, 1.9, and 7.1 kb. Recombination rates after one round of replication were 4.7, 7.4, and 8.2% with markers 1.0, 1.9, and 7.1 kb apart, respectively. Thus, the rate of homologous recombination with 1.0 kb of marker distance is similar in MLV and SNV. The recombination rate increases when the marker distance increases from 1.0 to 1.9 kb; however, the recombination rates with marker distances of 1.9 and 7.1 kb are not significantly different. These data refute the previous assumption that recombination is proportional to marker distance and define the maximum recombining population in retroviruses.
Figures




References
-
- Coffin J M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. - PubMed
-
- Coffin J M. Structure and classification of retroviruses. In: Levy J A, editor. The retroviridae. New York, N.Y: Plenum Press; 1992. pp. 19–49.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

