Use of differential display reverse transcription-PCR to reveal cellular changes during stimuli that result in herpes simplex virus type 1 reactivation from latency: upregulation of immediate-early cellular response genes TIS7, interferon, and interferon regulatory factor-1
- PMID: 9445025
- PMCID: PMC124603
- DOI: 10.1128/JVI.72.2.1252-1261.1998
Use of differential display reverse transcription-PCR to reveal cellular changes during stimuli that result in herpes simplex virus type 1 reactivation from latency: upregulation of immediate-early cellular response genes TIS7, interferon, and interferon regulatory factor-1
Abstract
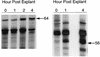
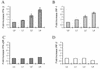
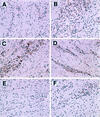
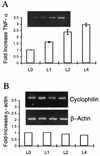
The detailed mechanism which governs the choice between herpes simplex virus (HSV) latency and reactivation remains to be elucidated. It is probable that altered expression of cellular factors in sensory neurons leads to induction of HSV gene expression resulting in reactivation. As an approach to identify novel cellular genes which are activated or repressed by stimuli that reactivate HSV from latency and hence may play a role in viral reactivation, RNA from explanted trigeminal ganglia (TG) was analyzed by differential display reverse transcription-PCR (DDRT-PCR). Nearly 50 cDNAs whose mRNA level was modified by the stress of explantation were isolated and sequenced. We present a listing of a spectrum of altered RNAs, including both known and unknown sequences. Five of those differentially displayed transcripts were identified as interferon-related murine TIS7 mRNA. These results were confirmed in both infected and uninfected ganglia by quantitative RNase protection assay and immunostaining. Alpha and beta interferons and interferon regulatory factor-1 (IRF-1) were also induced by explantation. In addition, we have identified sequences that correspond to IRF-1 consensus binding sites in both HSV type 1 origins of replication. Our findings suggest that physiological pathways that include these cellular factors may be involved in modulating HSV reactivation.
Figures




Similar articles
-
The herpes simplex virus type 1 ICP0 promoter is activated by viral reactivation stimuli in trigeminal ganglia neurons of transgenic mice.J Neurovirol. 2003 Jun;9(3):336-45. doi: 10.1080/13550280390201047. J Neurovirol. 2003. PMID: 12775417
-
Antagonizing the Glucocorticoid Receptor Impairs Explant-Induced Reactivation in Mice Latently Infected with Herpes Simplex Virus 1.J Virol. 2019 Jun 14;93(13):e00418-19. doi: 10.1128/JVI.00418-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30971470 Free PMC article.
-
Detection of herpes simplex virus type 1-encoded RNA by polymerase chain reaction: different pattern of viral RNA detection in latently infected murine trigeminal ganglia following in vitro or in vivo reactivation.J Gen Virol. 1994 Mar;75 ( Pt 3):647-50. doi: 10.1099/0022-1317-75-3-647. J Gen Virol. 1994. PMID: 8126462
-
A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review.
-
Early expression of herpes simplex virus (HSV) proteins and reactivation of latent infection.Folia Microbiol (Praha). 2000;45(1):7-28. doi: 10.1007/BF02817445. Folia Microbiol (Praha). 2000. PMID: 11200675 Review.
Cited by
-
Gene array analysis reveals changes in peripheral nervous system gene expression following stimuli that result in reactivation of latent herpes simplex virus type 1: induction of transcription factor Bcl-3.J Virol. 2001 Oct;75(20):9909-17. doi: 10.1128/JVI.75.20.9909-9917.2001. J Virol. 2001. PMID: 11559823 Free PMC article.
-
HSV-1-based vectors for gene therapy of neurological diseases and brain tumors: part I. HSV-1 structure, replication and pathogenesis.Neoplasia. 1999 Nov;1(5):387-401. doi: 10.1038/sj.neo.7900055. Neoplasia. 1999. PMID: 10933054 Free PMC article. Review.
-
The cellular response to herpes simplex virus type 1 (HSV-1) during latency and reactivation.J Neurovirol. 2005 Aug;11(4):376-83. doi: 10.1080/13550280591002405. J Neurovirol. 2005. PMID: 16162480
-
Vestibular Neuritis Following COVID-19 Vaccination: A Retrospective Study.Cureus. 2022 Apr 19;14(4):e24277. doi: 10.7759/cureus.24277. eCollection 2022 Apr. Cureus. 2022. PMID: 35602793 Free PMC article.
-
Adenovirus induction of an interferon-regulatory factor during entry into the late phase of infection.J Virol. 1998 Nov;72(11):9257-66. doi: 10.1128/JVI.72.11.9257-9266.1998. J Virol. 1998. PMID: 9765473 Free PMC article.
References
-
- Adams R L, Springall D R, Levene M M, Bushell T E. The immunocytochemical detection of herpes simplex virus in cervical smears—a valuable technique for routine use. J Pathol. 1984;143:241–247. - PubMed
-
- Altschul S F, Gisg W, Miller W, Meyers E W, Lipman D J. BLAST: a local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Arenalder A T, Lim R W, Varnum B C, Cole R, Vellis J D, Herschman H R. TIS gene expression in cultured rat astrocytes: induction by mitogens and stellation agents. J Neurosci Res. 1989;23:247–256. - PubMed
-
- Arvidson B. Retrograde axonal transport of horseradish peroxidase from cornea to trigeminal ganglion. Acta Neuropathol. 1977;38:49–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

