Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines
- PMID: 9445059
- PMCID: PMC124637
- DOI: 10.1128/JVI.72.2.1552-1576.1998
Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines
Abstract
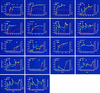
We have studied 18 participants in phase I/II clinical trials of recombinant gp120 (rgp120) subunit vaccines (MN and SF-2) who became infected with human immunodeficiency virus type 1 (HIV-1) during the course of the trials. Of the 18 individuals, 2 had received a placebo vaccine, 9 had been immunized with MN rgp120, and seven had been immunized with SF-2 rgp120. Thirteen of the 18 infected vaccinees had received three or four immunizations prior to becoming infected. Of these, two were placebo recipients, six had received MN rgp120, and five had received SF-2 rgp120. Only 1 of the 11 rgp120 recipients who had multiple immunizations failed to develop a strong immunoglobulin G antibody response to the immunogen. However, the antibody response to rgp120 was transient, typically having a half-life of 40 to 60 days. No significant neutralizing activity against the infecting strain was detected in any of the infected individuals at any time prior to infection. Antibody titers in subjects infected despite vaccination and in noninfected subjects were not significantly different. Envelope-specific cytotoxic T-lymphocyte responses measured after infection were infrequent and weak in the nine vaccinees who were tested. HIV-1 was isolated successfully from all 18 individuals. Sixteen of these strains had a non-syncytium-inducing (NSI) phenotype, while two had a syncytium-inducing (SI) phenotype. NSI strains used the CCR5 coreceptor to enter CD4+ cells, while an SI strain from one of the vaccinees also used CXCR4. Viruses isolated from the blood of rgp120 vaccinees were indistinguishable from viruses isolated from control individuals in terms of their inherent sensitivity to neutralization by specific monoclonal antibodies and their replication rates in vitro. Furthermore, genetic sequencing of the env genes of strains infecting the vaccinees did not reveal any features that clearly distinguished these viruses from contemporary clade B viruses circulating in the United States. Thus, despite rigorous genetic analyses, using various breakdowns of the data sets, we could find no evidence that rgp120 vaccination exerted selection pressure on the infecting HIV-1 strains. The viral burdens in the infected rgp120 vaccine recipients were also determined, and they were found to be not significantly different from those in cohorts of placebo-vaccinated and nonvaccinated individuals. In summary, we conclude that vaccination with rgp120 has had,to date, no obvious beneficial or adverse effects on the individuals we have studied.
Figures
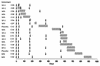






References
-
- Allaway G P, Davis-Bruno K L, Beaudry G A, Garcia E B, Wong E L, Ryder A M, Hasel K W, Gauduin M-C, Koup R A, McDougal J S, Maddon P J. Expression and characterization of CD4-IgG2, a novel heterotetramer which neutralizes primary HIV-1 isolates. AIDS Res Hum Retroviruses. 1995;11:533–540. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Artenstein A W, VanCott T C, Sitz K V, Robb M L, Wagner K F, Veit S C D, Rogers A F, Garner R P, Byron J W, Burnett P R, Birx D L. Mucosal immune responses in four distinct compartments of women infected with human immunodeficiency virus type 1: a comparison by site and correlation with clinical information. J Infect Dis. 1997;175:265–271. - PubMed
-
- Åsjö B, Morfeldt-Månson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyö E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;i:660–662. - PubMed
-
- Baenziger J, Liu Y, Walker C. Proceedings of the Seventh Annual Meeting of the National Cooperative Vaccine Development Groups for AIDS, Reston, Va. 1994. Cytotoxic T lymphocyte (CTL) responses in baboons and humans immunized with a recombinant HIV-1 gp120 vaccine; p. 88.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

