Differential expression of individual suppressor tRNA(Trp) gene gene family members in vitro and in vivo in the nematode Caenorhabditis elegans
- PMID: 9447966
- PMCID: PMC108781
- DOI: 10.1128/MCB.18.2.703
Differential expression of individual suppressor tRNA(Trp) gene gene family members in vitro and in vivo in the nematode Caenorhabditis elegans
Abstract
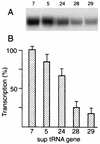
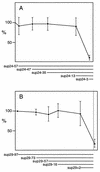
Eight different amber suppressor tRNA (suptRNA) mutations in the nematode Caenorhabditis elegans have been isolated; all are derived from members of the tRNA(Trp) gene family (K. Kondo, B. Makovec, R. H. Waterston, and J. Hodgkin, J. Mol. Biol. 215:7-19, 1990). Genetic assays of suppressor activity suggested that individual tRNA genes were differentially expressed, probably in a tissue- or developmental stage-specific manner. We have now examined the expression of representative members of this gene family both in vitro, using transcription in embryonic cell extracts, and in vivo, by assaying suppression of an amber-mutated lacZ reporter gene in animals carrying different suptRNA mutations. Individual wild-type tRNA(Trp) genes and their amber-suppressing counterparts appear to be transcribed and processed identically in vitro, suggesting that the behavior of suptRNAs should reflect wild-type tRNA expression. The levels of transcription of different suptRNA genes closely parallel the extent of genetic suppression in vivo. The results suggest that differential expression of tRNA genes is most likely at the transcriptional rather than the posttranscriptional level and that 5' flanking sequences play a role in vitro, and probably in vivo as well. Using suppression of a lacZ(Am) reporter gene as a more direct assay of suptRNA activity in individual cell types, we have again observed differential expression which correlates with genetic and in vitro transcription results. This provides a model system to more extensively study the basis for differential expression of this tRNA gene family.
Figures


References
-
- Candelas G C, Arroyo G, Carrasco C, Dompensciel R. Spider silkglands contain a tissue-specific alanine tRNA that accumulates in vitro in response to the stimulus for silk protein synthesis. Dev Biol. 1990;140:215–220. - PubMed
-
- Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. - PubMed
-
- Dingermann T, Amon-Bohm E, Bertling W, Marschalek R, Nerke K. A family of non-allelic tRNAGUUVal genes from the cellular slime mold Dictyostelium discoideum. Gene. 1988;73:373–384. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
