A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos
- PMID: 9447994
- PMCID: PMC108809
- DOI: 10.1128/MCB.18.2.967
A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos
Abstract
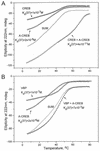
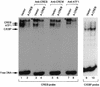
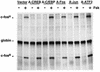
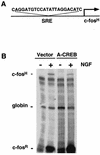
Several studies have characterized the upstream regulatory region of c-fos, and identified cis-acting elements termed the cyclic AMP (cAMP) response elements (CREs) that are critical for c-fos transcription in response to a variety of extracellular stimuli. Although several transcription factors can bind to CREs in vitro, the identity of the transcription factor(s) that activates the c-fos promoter via the CRE in vivo remains unclear. To help identify the trans-acting factors that regulate stimulus-dependent transcription of c-fos via the CREs, dominant-negative (D-N) inhibitor proteins that function by preventing DNA binding of B-ZIP proteins in a dimerization domain-dependent fashion were developed. A D-N inhibitor of CREB, termed A-CREB, was constructed by fusing a designed acidic amphipathic extension onto the N terminus of the CREB leucine zipper domain. The acidic extension of A-CREB interacts with the basic region of CREB forming a coiled-coil extension of the leucine zipper and thus prevents the basic region of wild-type CREB from binding to DNA. Other D-N inhibitors generated in a similar manner with the dimerization domains of Fos, Jun, C/EBP, ATF-2, or VBP did not block CREB DNA binding activity, nor did they inhibit transcriptional activation of a minimal promoter containing a single CRE in PC12 cells. A-CREB inhibited activation of CRE-mediated transcription evoked by three distinct stimuli: forskolin, which increases intracellular cAMP; membrane depolarization, which promotes Ca2+ influx; and nerve growth factor (NGF). A-CREB completely inhibited cAMP-mediated, but only partially inhibited Ca2+- and NGF-mediated, transcription of a reporter gene containing 750 bp of the native c-fos promoter. Moreover, glutamate induction of c-fos expression in primary cortical neurons was dependent on CREB. In contrast, induction of c-fos transcription by UV light was not inhibited by A-CREB. Lastly, A-CREB attenuated NGF induction of morphological differentiation in PC12 cells. These results suggest that CREB or its closely related family members are general mediators of stimulus-dependent transcription of c-fos and are required for at least some of the long-term actions of NGF.
Figures







References
-
- Bading H, Ginty D D, Greenberg M E. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathway. Science. 1993;260:181–186. - PubMed
-
- Barton H, Muthusamy M, Chanyangam C, Fischer C, Cledenin C, Leiden J M. Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature. 1996;379:81–85. - PubMed
-
- Blackstone C D, Moss S J, Martin L J, Price D L, Huganir R L. Biochemical characterization and localization of a non-NMDA receptor in rat brain. J Neurochem. 1992;58:1118–1126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
