The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein
- PMID: 9448006
- PMCID: PMC108821
- DOI: 10.1128/MCB.18.2.1084
The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein
Abstract
PML is a nuclear protein with growth-suppressive properties originally identified in the context of the PML-retinoic acid receptor alpha (RAR alpha) fusion protein of acute promyelocytic leukemia. PML localizes within distinct nuclear structures, called nuclear bodies, which are disrupted by the expression of PML-RAR alpha. We report that PML colocalizes with the nonphosphorylated fraction of the retinoblastoma protein (pRB) within nuclear bodies and that pRB is delocalized by PML-RAR alpha expression. Both PML and PML-RAR alpha form complexes with the nonphosphorylated form of pRB in vivo, and they interact with the pocket region of pRB. The regions of PML and PML-RAR alpha involved in pRB binding differ; in fact, the B boxes and the C-terminal region of PML, the latter of which is not present in PML-RAR alpha, are essential for the formation of stable complexes with pRB. Functionally, PML abolishes activation of glucocorticoid receptor-regulated transcription by pRB, whereas PML-RAR alpha further increases it. Our results suggest that PML may be part of transcription-regulatory complexes and that the oncogenic potential of the PML-RAR alpha protein may derive from the alteration of PML-regulated transcription.
Figures





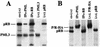
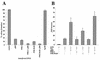
References
-
- Alcalay M, Zangrilli D, Pandolfi P P, Longo L, Mencarelli A, Giacomucci A, Rocchi M, Biondi A, Rambaldi A, Lo Coco F, Diverio D, Donti E, Grignani F, Pelicci P G. Translocation breakpoint of acute promyelocytic leukemia lies within the retinoic acid receptor alpha locus. Proc Natl Acad Sci USA. 1991;88:1977–1981. - PMC - PubMed
-
- Baker S J, Markowitz S, Fearon E R, Willson K V, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. - PubMed
-
- Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene. 1996;13:971–982. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
