Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo
- PMID: 9448009
- PMCID: PMC108824
- DOI: 10.1128/MCB.18.2.1115
Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo
Abstract
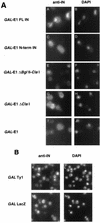
Retrotransposon Ty1 faces a formidable cell barrier during transposition--the yeast nuclear membrane which remains intact throughout the cell cycle. We investigated the mechanism by which transposition intermediates are transported from the cytoplasm (the presumed site of Ty1 DNA synthesis) to the nucleus, where they are integrated into the genome. Ty1 integrase has a nuclear localization signal (NLS) at its C terminus. Both full-length integrase and a C-terminal fragment localize to the nucleus. C-terminal deletion mutants in Ty1 integrase were used to map the putative NLS to the last 74 amino acid residues of integrase. Mutations in basic segments within this region decreased retrotransposition at least 50-fold in vivo. Furthermore, these mutant integrase proteins failed to localize to the nucleus. Production of virus-like particles, reverse transcriptase activity, and complete in vitro Ty1 integration resembled wild-type levels, consistent with failure of the mutant integrases to enter the nucleus.
Figures




References
-
- Adams A E, Pringle J R. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 1991;194:729–731. - PubMed
-
- Aitchison J D, Rout M P, Marelli M, Blobel G, Wozniak R W. Two novel related yeast nucleoporins, Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. - PMC - PubMed
-
- Bassett, D. Personal communication.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
