PCR analysis of tissue samples from the 1979 Sverdlovsk anthrax victims: the presence of multiple Bacillus anthracis strains in different victims
- PMID: 9448313
- PMCID: PMC18726
- DOI: 10.1073/pnas.95.3.1224
PCR analysis of tissue samples from the 1979 Sverdlovsk anthrax victims: the presence of multiple Bacillus anthracis strains in different victims
Abstract
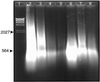
An outbreak of human anthrax occurred in Sverdlovsk, Union of Soviet Socialists Republic (now Ekaterinburg, Russia) in April 1979. Officials attributed this to consumption of contaminated meat, but Western governments believed it resulted from inhalation of spores accidentally released from a nearby military research facility. Tissue samples from 11 victims were obtained and methods of efficiently extracting high-quality total DNA from these samples were developed. Extracted DNA was analyzed by using PCR to determine whether it contained Bacillus anthracis-specific sequences. Double PCR using "nested primers" increased sensitivity of the assay significantly. Tissue samples from 11 persons who died during the epidemic were examined. Results demonstrated that the entire complement of B. anthracis toxin and capsular antigen genes required for pathogenicity were present in tissues from each of these victims. Tissue from a vaccination site contained primarily nucleic acids from a live vaccine, although traces of genes from the infecting organisms were also present. PCR analysis using primers that detect the vrrA gene variable region on the B. anthracis chromosome demonstrated that at least four of the five known strain categories defined by this region were present in the tissue samples. Only one category is found in a single B. anthracis strain.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

