A gene encoding antigenic peptides of human squamous cell carcinoma recognized by cytotoxic T lymphocytes
- PMID: 9449708
- PMCID: PMC2212124
- DOI: 10.1084/jem.187.3.277
A gene encoding antigenic peptides of human squamous cell carcinoma recognized by cytotoxic T lymphocytes
Abstract
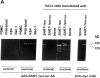
Except for melanomas, tumor antigens recognized by cytotoxic T lymphocytes (CTLs) are yet unidentified. We have identified a gene encoding antigenic peptides of human squamous cell carcinomas (SCCs) recognized by human histocompatibility leukocyte antigens (HLA)- A2601-restricted CTLs. This gene showed no similarity to known sequences, and encoded two (125- and 43-kilodalton [kD]) proteins. The 125-kD protein with the leucine zipper motif was expressed in the nucleus of the majority of proliferating cells tested, including normal and malignant cells. The 43-kD protein was expressed in the cytosol of most SCCs from various organs and half of lung adenocarcinomas, but was not expressed in other cancers nor in a panel of normal tissues. The three nonapeptides shared by the two proteins were recognized by the KE4 CTLs, and one of the peptides induced in vitro from peripheral blood mononuclear cells (PBMCs) the CTLs restricted to the autologous tumor cells. The 43-kD protein and this nonapeptide (KGSGKMKTE) may be useful for the specific immunotherapy of HLA-A2601(+) epithelial cancer patients.
Figures
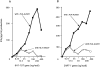








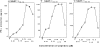




Similar articles
-
Identification of a gene coding for a new squamous cell carcinoma antigen recognized by the CTL.J Immunol. 2000 Mar 1;164(5):2565-74. doi: 10.4049/jimmunol.164.5.2565. J Immunol. 2000. PMID: 10679095
-
Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients.Cancer Res. 1999 Aug 15;59(16):4056-63. Cancer Res. 1999. PMID: 10463607
-
Induction of human leukocyte antigen-A26-restricted and tumor-specific cytotoxic T lymphocytes by a single peptide of the SART1 antigen in patients with cancer with different A26 subtypes.J Immunother. 2000 May-Jun;23(3):296-303. doi: 10.1097/00002371-200005000-00002. J Immunother. 2000. PMID: 10838658
-
[Tumor-rejection antigens expressed on human squamous cell carcinoma].Hum Cell. 1995 Dec;8(4):149-54. Hum Cell. 1995. PMID: 8721083 Review. Japanese.
-
Identification of genes coding for tumor antigens recognized by cytolytic T lymphocytes.Methods. 1997 Jun;12(2):125-42. doi: 10.1006/meth.1997.0462. Methods. 1997. PMID: 9184377 Review.
Cited by
-
[Squamous cell carcinoma of the head and neck. Principles and current concepts of immunotherapy].HNO. 2005 Mar;53(3):285-97; quiz 298. doi: 10.1007/s00106-004-1167-0. HNO. 2005. PMID: 15759168 Review. German.
-
A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia.J Exp Med. 1999 Jan 18;189(2):301-8. doi: 10.1084/jem.189.2.301. J Exp Med. 1999. PMID: 9892612 Free PMC article.
-
Expression of the SART-1 antigens in uterine cancers.Jpn J Cancer Res. 1998 Dec;89(12):1292-5. doi: 10.1111/j.1349-7006.1998.tb00526.x. Jpn J Cancer Res. 1998. PMID: 10081490 Free PMC article.
-
Successful induction of tumor-specific cytotoxic T lymphocytes from patients with non-small cell lung cancer using CD80-transfected autologous tumor cells.Jpn J Cancer Res. 2001 Mar;92(3):309-15. doi: 10.1111/j.1349-7006.2001.tb01096.x. Jpn J Cancer Res. 2001. PMID: 11267941 Free PMC article.
-
Therapeutic Cancer Vaccines-Antigen Discovery and Adjuvant Delivery Platforms.Pharmaceutics. 2022 Jul 11;14(7):1448. doi: 10.3390/pharmaceutics14071448. Pharmaceutics. 2022. PMID: 35890342 Free PMC article. Review.
References
-
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. - PubMed
-
- Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, Rosenberg SA. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivotumor rejection. Proc Natl Acad Sci USA. 1994;91:6458–6462. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

