Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa
- PMID: 9449836
- PMCID: PMC35188
- DOI: 10.1104/pp.116.1.53
Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa
Abstract
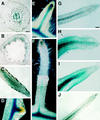
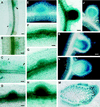
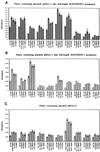
ENOD40, an early nodulin gene, is expressed following inoculation with Rhizobium meliloti or by adding R. meliloti-produced nodulation (Nod) factors or the plant hormone cytokinin to uninoculated roots. We isolated two MsENOD40 clones, designated MsENOD40-1 and MsENOD40-2, with distinct promoters from an alfalfa (Medicago sativa cv Chief) genomic library. The promoters were fused to the reporter gene uidA (gus), and the constructs were introduced into alfalfa. We observed that the MsENOD40-1 construct was expressed almost exclusively under symbiotic conditions. The MsENOD40-2 construct was transcribed under both symbiotic and nonsymbiotic conditions and in nonnodular and nodular tissues. Both MsENOD40 promoter-gus constructs were similarly expressed as nodules developed, and both were expressed in roots treated with 6-benzylaminopurine or purified Nod factor. However, no blue color was detected in nodule-like structures induced by the auxin transport inhibitor N-1-(naphthyl)phthalamic acid on roots of plants containing the MsENOD40-1 promoter construct, whereas pseudonodules from plants containing the MsENOD40-2 promoter construct stained blue. A 616-bp region at the distal 5' end of the promoter is important for proper spatial expression of MsENOD40 in nodules and also for Nod-factor and cytokinin-induced expression.
Figures






References
-
- Asad S, Fang Y, Wycoff KL, Hirsch AM. Isolation and characterization of cDNA and genomic clones of MsENOD40: transcripts are detected in meristematic cells of alfalfa. Protoplasma. 1994;183:10–23.
-
- Bak Ramlov K, Bech Laursen N, Stougaard J, Marcker KA. Site-directed mutagenesis of the organ-specific element in the soybean leghaemobloin lbc3 gene promoter. Plant J. 1993;4:577–580. - PubMed
-
- Bauer P, Ratet P, Crespi MD, Schultze M, Kondorosi A. Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and Msenod12A expression in alfalfa roots. Plant J. 1996;10:91–105.
-
- Beaty JS, Powell GK, Licia L, Rigier DA, MacDonald EMS, Hommes NG, Morris RO. Tzs, a nopaline Ti plasmid gene from Agrobacterium tumefaciens associated with trans-zeatin biosynthesis. Mol Gen Genet. 1986;203:274–280.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

