Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus
- PMID: 9449838
- PMCID: PMC35191
- DOI: 10.1104/pp.116.1.91
Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus
Abstract
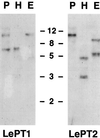
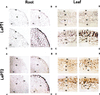
Phosphorus is a major nutrient acquired by roots via high-affinity inorganic phosphate (Pi) transporters. In this paper, we describe the tissue-specific regulation of tomato (Lycopersicon esculentum L.) Pi-transporter genes by Pi. The encoded peptides of the LePT1 and LePT2 genes belong to a family of 12 membrane-spanning domain proteins and show a high degree of sequence identity to known high-affinity Pi transporters. Both genes are highly expressed in roots, although there is some expression of LePT1 in leaves. Their expression is markedly induced by Pi starvation but not by starvation of nitrogen, potassium, or iron. The transcripts are primarily localized in root epidermis under Pi starvation. Accumulation of LePT1 message was also observed in palisade parenchyma cells of Pi-starved leaves. Our data suggest that the epidermally localized Pi transporters may play a significant role in acquiring the nutrient under natural conditions. Divided root-system studies support the hypothesis that signal(s) for the Pi-starvation response may arise internally because of the changes in cellular concentration of phosphorus.
Figures







References
-
- Anghinoni I, Barber SA. Phosphorus influx and growth characteristics of corn roots as influenced by phosphorus supply. Agron J. 1980;72:685–688.
-
- Barber SA, Walker JM, Vasey EH. Mechanisms for the movement of plant nutrients from the soil and fertilizer to the plant root. J Agric Food Chem. 1963;11:204–207.
-
- Bariola PA, Howard CJ, Taylor CP, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to Pi limitation. Plant J. 1994;6:673–685. - PubMed
-
- Clarkson DT (1991) Root structure and sites of ion uptake. In WY Waisel, A Eshel, U Kafkafi, eds, Plant Roots: The Hidden Half. M. Dekker, New York, pp 417–453
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

