Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation
- PMID: 9449848
- PMCID: PMC35176
- DOI: 10.1104/pp.116.1.357
Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation
Abstract
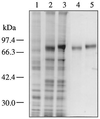
We have investigated two NADPH-cytochrome (Cyt) P450 reductase isoforms encoded by separate genes (AR1 and AR2) in Arabidopsis thaliana. We isolated AR1 and AR2 cDNAs using a mung bean (Phaseolus aureus L.) NADPH-Cyt P450 reductase cDNA as a probe. The recombinant AR1 and AR2 proteins produced using a baculovirus expression system showed similar Km values for Cyt c and NADPH, respectively. In the reconstitution system with a recombinant cinnamate 4-hydroxylase (CYP73A5), the recombinant AR1 and AR2 proteins gave the same level of cinnamate 4-hydroxylase activity (about 70 nmol min-1 nmol-1 P450). The AR2 gene expression was transiently induced by 4- and 3-fold within 1 h of wounding and light treatments, respectively, and the induction time course preceded those of CYP73A5 and a phenylalanine ammonia-lyase (PAL1) gene. On the contrary, the AR1 expression level did not change during the treatments. Analysis of the AR1 and AR2 gene structure revealed that only the AR2 promoter contained three putative sequence motifs (boxes P, A, and L), which are involved in the coordinated expression of CYP73A5 and other phenylpropanoid pathway genes. These results suggest the possibility that AR2 transcription may be functionally linked to the induced levels of phenylpropanoid pathway enzymes.
Figures


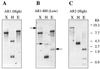



References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Siedman JG, Smith JA, Struhl K (1987) Current Protocols in Molecular Biology. John Wiley, New York
-
- Benveniste I, Lesot A, Hasenfratz MP, Kochs G, Durst F. Multiple forms of NADPH-cytochrome P450 reductase in higher plants. Biochem Biophys Res Commun. 1991;177:105–112. - PubMed
-
- Bolwell GP, Bozak K, Zimmerlin A. Plant cytochrome P450. Phytochemistry. 1994;37:1491–1506. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

