Motivation and organizational principles for anatomical knowledge representation: the digital anatomist symbolic knowledge base
- PMID: 9452983
- PMCID: PMC61273
- DOI: 10.1136/jamia.1998.0050017
Motivation and organizational principles for anatomical knowledge representation: the digital anatomist symbolic knowledge base
Abstract
Objective: Conceptualization of the physical objects and spaces that constitute the human body at the macroscopic level of organization, specified as a machine-parseable ontology that, in its human-readable form, is comprehensible to both expert and novice users of anatomical information.
Design: Conceived as an anatomical enhancement of the UMLS Semantic Network and Metathesaurus, the anatomical ontology was formulated by specifying defining attributes and differentia for classes and subclasses of physical anatomical entities based on their partitive and spatial relationships. The validity of the classification was assessed by instantiating the ontology for the thorax. Several transitive relationships were used for symbolically modeling aspects of the physical organization of the thorax.
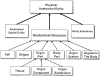
Results: By declaring Organ as the macroscopic organizational unit of the body, and defining the entities that constitute organs and higher level entities constituted by organs, all anatomical entities could be assigned to one of three top level classes (Anatomical structure, Anatomical spatial entity and Body substance). The ontology accommodates both the systemic and regional (topographical) views of anatomy, as well as diverse clinical naming conventions of anatomical entities.
Conclusions: The ontology formulated for the thorax is extendible to microscopic and cellular levels, as well as to other body parts, in that its classes subsume essentially all anatomical entities that constitute the body. Explicit definitions of these entities and their relationships provide the first requirement for standards in anatomical concept representation. Conceived from an anatomical viewpoint, the ontology can be generalized and mapped to other biomedical domains and problem solving tasks that require anatomical knowledge.
Figures




References
-
- National Library of Medicine. UMLS knowledge sources, 7th ed. Unified Medical Language System. Bethesda, MD: National Library of Medicine, 1996.
-
- Côté RA, Rothwell DJ, Palotay JL, Beckett RS, Brochu L (eds). SNOMED International: The systematized nomenclature of human and veterinary medicine. Northfield, IL: College of American Pathologists, 1993.
-
- Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modeling language for medical terminology. Artif Intell Med. 1997;9: 139-71. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

