Lactoferrin inhibits the endotoxin interaction with CD14 by competition with the lipopolysaccharide-binding protein
- PMID: 9453600
- PMCID: PMC107932
- DOI: 10.1128/IAI.66.2.486-491.1998
Lactoferrin inhibits the endotoxin interaction with CD14 by competition with the lipopolysaccharide-binding protein
Abstract
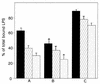
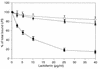
Human lactoferrin (hLf), a glycoprotein released from neutrophil granules during inflammation, and the lipopolysaccharide (LPS)-binding protein (LBP), an acute-phase serum protein, are known to bind to the lipid A of LPS. The LPS-binding sites are located in the N-terminal regions of both proteins, at amino acid residues 28 to 34 of hLf and 91 to 108 of LBP. Both of these proteins modulate endotoxin activities, but they possess biologically antagonistic properties. In this study, we have investigated the competition between hLf and recombinant human LBP (rhLBP) for the binding of Escherichia coli 055:B5 LPS to the differentiated monocytic THP-1 cell line. Our studies revealed that hLf prevented the rhLBP-mediated binding of LPS to the CD14 receptor on cells. Maximal inhibition of LPS-cell interactions by hLf was raised when both hLf and rhLBP were simultaneously added to LPS or when hLf and LPS were mixed with cells 30 min prior to the incubation with rhLBP. However, when hLf was added 30 min after the interaction of rhLBP with LPS, the binding of the rhLPS-LBP complex to CD14 could not be reversed. These observations indicate that hLf competes with rhLBP for the LPS binding and therefore interferes with the interaction of LPS with CD14. Furthermore, experiments involving competitive binding of the rhLBP-LPS complex to cells with two recombinant mutated hLfs show that in addition to residues 28 to 34, another basic cluster which contains residues 1 to 5 of hLf competes for the binding to LPS. Basic sequences homologous to residues 28 to 34 of hLf were evidenced on LPS-binding proteins such as LBP, bactericidal/permeability-increasing protein, and Limulus anti-LPS factor.
Figures





Similar articles
-
Lactoferrin-lipopolysaccharide interaction: involvement of the 28-34 loop region of human lactoferrin in the high-affinity binding to Escherichia coli 055B5 lipopolysaccharide.Biochem J. 1995 Dec 15;312 ( Pt 3)(Pt 3):839-45. doi: 10.1042/bj3120839. Biochem J. 1995. PMID: 8554529 Free PMC article.
-
3LPS-binding protein and its interactions with P. gingivalis LPS modulate pro-inflammatory response and Toll-like receptor signaling in human oral keratinocytes.PLoS One. 2017 Apr 6;12(4):e0173223. doi: 10.1371/journal.pone.0173223. eCollection 2017. PLoS One. 2017. PMID: 28384159 Free PMC article.
-
Human lactoferrin interacts with soluble CD14 and inhibits expression of endothelial adhesion molecules, E-selectin and ICAM-1, induced by the CD14-lipopolysaccharide complex.Infect Immun. 2000 Dec;68(12):6519-25. doi: 10.1128/IAI.68.12.6519-6525.2000. Infect Immun. 2000. PMID: 11083760 Free PMC article.
-
Function of lipopolysaccharide (LPS)-binding protein (LBP) and CD14, the receptor for LPS/LBP complexes: a short review.Res Immunol. 1992 Jan;143(1):11-5. doi: 10.1016/0923-2494(92)80074-u. Res Immunol. 1992. PMID: 1373512 Review.
-
LPS-binding proteins and receptors.J Leukoc Biol. 1998 Jul;64(1):25-32. doi: 10.1002/jlb.64.1.25. J Leukoc Biol. 1998. PMID: 9665271 Review.
Cited by
-
Lactoferrin protects rabbits from Shigella flexneri-induced inflammatory enteritis.Infect Immun. 2002 Dec;70(12):7050-3. doi: 10.1128/IAI.70.12.7050-7053.2002. Infect Immun. 2002. PMID: 12438385 Free PMC article.
-
Reciprocal interactions between lactoferrin and bacterial endotoxins and their role in the regulation of the immune response.Toxins (Basel). 2010 Jan;2(1):54-68. doi: 10.3390/toxins2010054. Epub 2010 Jan 8. Toxins (Basel). 2010. PMID: 22069546 Free PMC article. Review.
-
Establishment of a mouse model of lipopolysaccharide-induced neutrophilic nasal polyps.Exp Ther Med. 2017 Dec;14(6):5275-5282. doi: 10.3892/etm.2017.5208. Epub 2017 Sep 27. Exp Ther Med. 2017. PMID: 29285053 Free PMC article.
-
Safety and tolerability of the antimicrobial peptide human lactoferrin 1-11 (hLF1-11).BMC Med. 2009 Sep 8;7:44. doi: 10.1186/1741-7015-7-44. BMC Med. 2009. PMID: 19735580 Free PMC article. Clinical Trial.
-
Inhibition of DAMP actions in the tumoral microenvironment using lactoferrin-glycyrrhizin conjugate for glioblastoma therapy.Biomater Res. 2023 May 20;27(1):52. doi: 10.1186/s40824-023-00391-w. Biomater Res. 2023. PMID: 37210579 Free PMC article.
References
-
- Abrahamson S L, Wu H S, Williams R E, Der K, Ottah N, Little R, Gazzano-Santoro H, Theofan G, Bauer R, Leigh S, Orme A, Horwitz A H, Caroll S F, Dedrick R L. Biochemical characterization of recombinant fusions of LPS-binding protein and bactericidal/permeability-increasing protein. J Biol Chem. 1997;272:2149–2155. - PubMed
-
- Anderson B F, Baker H M, Norris G E, Rice D W, Baker E N. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 Å resolution. J Mol Biol. 1989;209:711–734. - PubMed
-
- Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K, Tomita M. Identification of the bactericidal domain of lactoferrin. Biochim Biophys Acta. 1992;1121:130–136. - PubMed
-
- Brock J. Iron and cells of the immune system. In: de Souza M, Brock J, editors. Iron in immunity, cancer and inflammation. Chichester, England: John Wiley and Sons; 1986. pp. 81–98.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

