Identification and characterization of the PutP proline permease that contributes to in vivo survival of Staphylococcus aureus in animal models
- PMID: 9453610
- PMCID: PMC107942
- DOI: 10.1128/IAI.66.2.567-572.1998
Identification and characterization of the PutP proline permease that contributes to in vivo survival of Staphylococcus aureus in animal models
Abstract
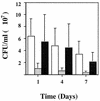
Staphylococcus aureus is an important pathogen of humans and other animals, causing bacteremia, abscesses, endocarditis, and other infectious syndromes. A signature-tagged mutagenesis (STM) system was adapted for use in studying the genes required for in vivo survival of S. aureus. An STM library was ultimately created in S. aureus RN6390, with Tn917 being used to create the transposon mutations. Pools of S. aureus RN6390 mutants were screened in mouse abscess, bacteremia, and wound infection models for growth attenuation after in vivo passage. One of the mutants that was identified displayed marked attenuation following large-pool screening in all three animal models, which was confirmed in bacteremia and endocarditis models of infection with a smaller pool of mutants. Sequence analysis of the entire open reading frame showed a 99% identity to the high-affinity proline permease (putP) gene characterized in another strain of S. aureus. In wound and murine abscess infection models, the putP mutant was approximately 10-fold more attenuated than was wild-type strain RN6390. Another S. aureus strain transduced with the putP mutation also displayed an attenuated phenotype after passage in the wound model. A [3H]proline uptake assay showed that less proline was specifically transported into the putP mutant than into strain RN6390. The reduced viability of the bacteria possessing the mutation in the S. aureus high-affinity proline permease suggests that proline scavenging by the bacteria is important for in vivo growth and proliferation and that analogs of proline may serve as potential antistaphylococcal therapeutic agents.
Figures

Similar articles
-
Low-proline environments impair growth, proline transport and in vivo survival of Staphylococcus aureus strain-specific putP mutants.Microbiology (Reading). 2004 Apr;150(Pt 4):1055-1061. doi: 10.1099/mic.0.26710-0. Microbiology (Reading). 2004. PMID: 15073314
-
Impact of the high-affinity proline permease gene (putP) on the virulence of Staphylococcus aureus in experimental endocarditis.Infect Immun. 1999 Feb;67(2):740-4. doi: 10.1128/IAI.67.2.740-744.1999. Infect Immun. 1999. PMID: 9916085 Free PMC article.
-
Proline transport in Salmonella typhimurium: putP permease mutants with altered substrate specificity.J Bacteriol. 1986 Nov;168(2):590-4. doi: 10.1128/jb.168.2.590-594.1986. J Bacteriol. 1986. PMID: 3536852 Free PMC article.
-
Transcriptional activation of the Staphylococcus aureus putP gene by low-proline-high osmotic conditions and during infection of murine and human tissues.Infect Immun. 2006 Jan;74(1):399-409. doi: 10.1128/IAI.74.1.399-409.2006. Infect Immun. 2006. PMID: 16368996 Free PMC article.
-
Topology and function of the Na+/proline transporter of Escherichia coli, a member of the Na+/solute cotransporter family.Biochim Biophys Acta. 1998 Jun 10;1365(1-2):60-4. doi: 10.1016/s0005-2728(98)00044-9. Biochim Biophys Acta. 1998. PMID: 9693722 Review.
Cited by
-
Discovery of the Membrane Binding Domain in Trifunctional Proline Utilization A.Biochemistry. 2017 Nov 28;56(47):6292-6303. doi: 10.1021/acs.biochem.7b01008. Epub 2017 Nov 15. Biochemistry. 2017. PMID: 29090935 Free PMC article.
-
Study of Staphylococcus aureus pathogenic genes by transfer and expression in the less virulent organism Streptococcus gordonii.Infect Immun. 2001 Feb;69(2):657-64. doi: 10.1128/IAI.69.2.657-664.2001. Infect Immun. 2001. PMID: 11159952 Free PMC article.
-
mSphere of Influence: Metabolic redundancies enhance pathogenesis.mSphere. 2024 Jul 30;9(7):e0023924. doi: 10.1128/msphere.00239-24. Epub 2024 Jul 3. mSphere. 2024. PMID: 38958458 Free PMC article.
-
Tricarboxylic acid cycle-dependent attenuation of Staphylococcus aureus in vivo virulence by selective inhibition of amino acid transport.Infect Immun. 2009 Oct;77(10):4256-64. doi: 10.1128/IAI.00195-09. Epub 2009 Aug 10. Infect Immun. 2009. PMID: 19667045 Free PMC article.
-
Proline transporters ProT and PutP are required for Staphylococcus aureus infection.PLoS Pathog. 2023 Jan 18;19(1):e1011098. doi: 10.1371/journal.ppat.1011098. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36652494 Free PMC article.
References
-
- Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bergdoll M S. Staphylococcus aureus. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 463–523.
-
- Berman H M, McGandy E L, Burgner II J W, VanEtten R L. The crystal and molecular structure of l-azetidine-2-carboxylic acid. A naturally occurring homolog of proline. J Am Chem Soc. 1969;91:6177–6182. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

