Identification of a putative precursor to the major surface glycoprotein of Pneumocystis carinii
- PMID: 9453635
- PMCID: PMC113502
- DOI: 10.1128/IAI.66.2.741-746.1998
Identification of a putative precursor to the major surface glycoprotein of Pneumocystis carinii
Abstract
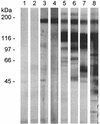
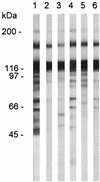
The major surface glycoprotein (MSG) of Pneumocystis carinii f. sp. carinii is a family of proteins encoded by a family of heterogeneous genes. Messenger RNAs encoding different MSGs each begin with the same 365-bp sequence, called the Upstream Conserved Sequence (UCS), which is in frame with the contiguous MSG sequence. The UCS contains several potential start sites for translation. To determine if translation of MSG mRNAs begins in the UCS, polyclonal antiserum was raised against the 123-amino-acid peptide encoded by the UCS. The anti-UCS serum reacted with a P. carinii protein that migrated at 170 kDa; however, it did not react with the mature MSG protein, which migrates at 116 kDa. A 170-kDa protein was immunoprecipitated with anti-UCS serum and shown to react with a monoclonal antibody against a conserved MSG epitope. To explore the functional role of the UCS in the trafficking of MSG, the nucleotide sequence encoding the UCS peptide was ligated to the 5' end of an MSG gene and incorporated into a recombinant baculovirus. Insect cells infected with the UCS-MSG hybrid gene expressed a 160-kDa protein which was N-glycosylated. By contrast, insect cells infected with a baculovirus carrying an MSG gene lacking the UCS expressed a nonglycosylated 130-kDa protein. These data suggest that in P. carinii, translation begins in the UCS to produce a pre-MSG protein, which is subsequently directed to the endoplasmic reticulum and processed to the mature form by proteolytic cleavage.
Figures



References
-
- Dei-Cas E, Mazars E, Ferragut C O, Durand I, Aliouat E-M, Dridba M, Palluault F, Cailliez J-C, Seguy N, Tibayrenc M, Mullet C, Creusy C, Camus D. Ultrastructural, genomic, isoenzymatic and biological features make it possible to distinguish rabbit Pneumocystis from other mammal Pneumocystis strains. J Eukaryot Microbiol. 1994;41:84S. - PubMed
-
- De Luca A, Ortona E, Margutti P, Visconti E, Tamburrini E, Siracusano A. Different amplification efficiency and nucleotide sequence variation in various Pneumocystis isolates from humans and rats. J Eukaryot Microbiol. 1994;41:85S. - PubMed
-
- Edlind T D, Bartlett M S, Weinberg G A, Prah G N, Smith J W. The beta-tubulin gene from rat and human isolates of Pneumocystis carinii. Mol Microbiol. 1992;6:3365–3373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

