Expression, structure, and location of epitopes of the major surface glycoprotein of Pneumocystis carinii f. sp. carinii
- PMID: 9455880
- PMCID: PMC121391
- DOI: 10.1128/CDLI.5.1.50-57.1998
Expression, structure, and location of epitopes of the major surface glycoprotein of Pneumocystis carinii f. sp. carinii
Abstract
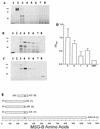
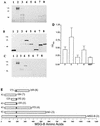
The major surface glycoprotein (MSG) of Pneumocystis carinii f. sp. carinii consists of a heterogeneous family of proteins that are encoded by approximately 100 unique genes. A genomic expression library was screened with a panel of MSG-specific monoclonal antibodies (MAbs) to identify conserved and rare epitopes. All of the antibodies reacted with epitopes that are encoded within the 5' end of MSG. The results from the expression screening identified antibodies that recognize highly conserved, moderately conserved, and rare epitopes. Four MAbs (MAbs RA-F1, RA-E7, RA-G10, and RB-E3) reacted with a maltose binding protein-MSG-B fusion protein ([MBP]MSG-B41-1065) by immunoblotting and enzyme-linked immunosorbent assay. Three of the MAbs (MAbs RA-F1, RA-G10, and RA-E7) reacted with the same continuous epitope that was localized to amino acids 278 to 290 of MSG-B. Comparison of the sequence of the RA-F1-, RA-G10-, and RA-E7-reactive epitope to the deduced amino acid sequences of multiple MSGs demonstrated that it is highly conserved. The reactivity of RB-E3 with MSG-B was shown to be dependent on amino acids 184 to 192, which may comprise a portion of a discontinuous epitope.
Figures


References
-
- Ausubel F M, Brent B, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1991.
-
- Barlow D J, Edwards M S, Thornton J M. Continuous and discontinuous protein antigenic determinants. Nature. 1986;322:747–748. - PubMed
-
- Cushion M T, Ruffolo J J, Walzer P D. Analysis of the developmental stages of Pneumocystis carinii, in vitro. Lab Invest. 1988;58:324–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

