Two roles for the DNA recognition site of the Klebsiella aerogenes nitrogen assimilation control protein
- PMID: 9457860
- PMCID: PMC106924
- DOI: 10.1128/JB.180.3.578-585.1998
Two roles for the DNA recognition site of the Klebsiella aerogenes nitrogen assimilation control protein
Abstract
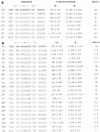
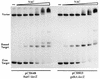
The nitrogen assimilation control protein (NAC) binds to a site within the promoter region of the histidine utilization operon (hutUH) of Klebsiella aerogenes, and NAC bound at this site activates transcription of hutUH. This NAC-binding site was characterized by a combination of random and directed DNA mutagenesis. Mutations that abolished or diminished in vivo transcriptional activation by NAC were found to lie within a 15-bp region contained within the 26-bp region protected by NAC from DNase I digestion. This 15-bp core has the palindromic ends ATA and TAT, and it matches the consensus for LysR family transcriptional regulators. Protein-binding experiments showed that transcriptional activation in vivo decreased with decreasing binding in vitro. In contrast to the NAC-binding site from hutUH, the NAC-binding site from the gdhA promoter failed to activate transcription from a semisynthetic promoter, and this failure was not due to weak binding or greatly distorted protein-DNA structure. Mutations in the promoter-proximal half-site of the NAC-binding site from gdhA allowed this site to activate transcription. Similar studies using the NAC-binding site from hut showed that two mutations in the promoter proximal half-site increased binding but abolished transcriptional activation. Interestingly, for symmetric mutations in the promoter-distal half-site, loss of transcriptional activation was always correlated with a decrease in binding. We conclude from these observations that if the binding in vitro reflects the binding in vivo, then binding of NAC to DNA is not sufficient for transcriptional activation and that the NAC-binding site can be functionally divided in two half-sites, with related but different functions.
Figures

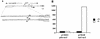




Similar articles
-
A NAC for regulating metabolism: the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae.J Bacteriol. 2010 Oct;192(19):4801-11. doi: 10.1128/JB.00266-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675498 Free PMC article. Review.
-
Importance of tetramer formation by the nitrogen assimilation control protein for strong repression of glutamate dehydrogenase formation in Klebsiella pneumoniae.J Bacteriol. 2005 Dec;187(24):8291-9. doi: 10.1128/JB.187.24.8291-8299.2005. J Bacteriol. 2005. PMID: 16321933 Free PMC article.
-
The amino-terminal 100 residues of the nitrogen assimilation control protein (NAC) encode all known properties of NAC from Klebsiella aerogenes and Escherichia coli.J Bacteriol. 1999 Feb;181(3):934-40. doi: 10.1128/JB.181.3.934-940.1999. J Bacteriol. 1999. PMID: 9922258 Free PMC article.
-
Bidirectional promoter in the hut(P) region of the histidine utilization (hut) operons from Klebsiella aerogenes.J Bacteriol. 1988 May;170(5):2240-6. doi: 10.1128/jb.170.5.2240-2246.1988. J Bacteriol. 1988. PMID: 2834335 Free PMC article.
-
The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes.Mol Microbiol. 1991 Nov;5(11):2575-80. doi: 10.1111/j.1365-2958.1991.tb01965.x. Mol Microbiol. 1991. PMID: 1664020 Review.
Cited by
-
Model-driven experimental design workflow expands understanding of regulatory role of Nac in Escherichia coli.NAR Genom Bioinform. 2023 Jan 20;5(1):lqad006. doi: 10.1093/nargab/lqad006. eCollection 2023 Mar. NAR Genom Bioinform. 2023. PMID: 36685725 Free PMC article.
-
A NAC for regulating metabolism: the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae.J Bacteriol. 2010 Oct;192(19):4801-11. doi: 10.1128/JB.00266-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675498 Free PMC article. Review.
-
Importance of tetramer formation by the nitrogen assimilation control protein for strong repression of glutamate dehydrogenase formation in Klebsiella pneumoniae.J Bacteriol. 2005 Dec;187(24):8291-9. doi: 10.1128/JB.187.24.8291-8299.2005. J Bacteriol. 2005. PMID: 16321933 Free PMC article.
-
Properties of the NAC (nitrogen assimilation control protein)-binding site within the ureD promoter of Klebsiella pneumoniae.J Bacteriol. 2010 Oct;192(19):4821-6. doi: 10.1128/JB.00883-09. Epub 2010 Jul 9. J Bacteriol. 2010. PMID: 20622063 Free PMC article.
-
Regulation of the histidine utilization (hut) system in bacteria.Microbiol Mol Biol Rev. 2012 Sep;76(3):565-84. doi: 10.1128/MMBR.00014-12. Microbiol Mol Biol Rev. 2012. PMID: 22933560 Free PMC article. Review.
References
-
- Bender R A. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol Microbiol. 1991;5:2575–2580. - PubMed
-
- Brenowitz M, Senear D. DNase I footprint analysis of protein-DNA binding. In: Ausubel F R, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1992. pp. 12.4.1–12.4.16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

