Modulation of the function of the signal receptor domain of XylR, a member of a family of prokaryotic enhancer-like positive regulators
- PMID: 9457863
- PMCID: PMC106927
- DOI: 10.1128/JB.180.3.600-604.1998
Modulation of the function of the signal receptor domain of XylR, a member of a family of prokaryotic enhancer-like positive regulators
Abstract
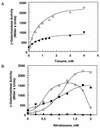
The XylR protein controls expression from the Pseudomonas putida TOL plasmid upper pathway operon promoter (Pu) in response to aromatic effectors. XylR-dependent stimulation of transcription from a Pu::lacZ fusion shows different induction kinetics with different effectors. With toluene, activation followed a hyperbolic curve with an apparent K of 0.95 mM and a maximum beta-galactosidase activity of 2,550 Miller units. With o-nitrotoluene, in contrast, activation followed a sigmoidal curve with an apparent K of 0.55 mM and a Hill coefficient of 2.65. m-Nitrotoluene kept the XylR regulator in an inactive transcriptional form. Therefore, upon binding of an effector, the substituent on the aromatic ring leads to productive or unproductive XylR forms. The different transcriptional states of the XylR regulator are substantiated by XylR mutants. XylRE172K is a mutant regulator that is able to stimulate transcription from the Pu promoter in the presence of m-nitrotoluene; however, its response to m-aminotoluene was negligible, in contrast with the wild-type regulator. These results illustrate the importance of the electrostatic interactions in effector recognition and in the stabilization of productive and unproductive forms by the regulator upon aromatic binding. XylRD135N and XylRD135Q are mutant regulators that are able to stimulate transcription from Pu in the absence of effectors, whereas substitution of Glu for Asp135 in XylRD135E resulted in a mutant whose ability to recognize effectors was severely impaired. Therefore, the conformation of mutant XylRD135Q as well as XylRD135N seemed to mimic that of the wild-type regulator when effector binding occurred, whereas mutant XylRD135E seemed to be blocked in a conformation similar to that of wild-type XylR and XylRE172K upon binding to an inhibitor molecule such as m-nitrotoluene or m-aminotoluene.
Figures


Similar articles
-
Single amino acids changes in the signal receptor domain of XylR resulted in mutants that stimulate transcription in the absence of effectors.J Biol Chem. 1995 Mar 10;270(10):5144-50. doi: 10.1074/jbc.270.10.5144. J Biol Chem. 1995. PMID: 7890623
-
In vitro activities of an N-terminal truncated form of XylR, a sigma 54-dependent transcriptional activator of Pseudomonas putida.J Mol Biol. 1996 May 17;258(4):575-87. doi: 10.1006/jmbi.1996.0270. J Mol Biol. 1996. PMID: 8636993
-
Genetic evidence for activation of the positive transcriptional regulator Xy1R, a member of the NtrC family of regulators, by effector binding.J Biol Chem. 1994 Mar 18;269(11):8059-62. J Biol Chem. 1994. PMID: 8132529
-
Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators.Annu Rev Microbiol. 1997;51:341-73. doi: 10.1146/annurev.micro.51.1.341. Annu Rev Microbiol. 1997. PMID: 9343354 Review.
-
Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways.Mol Microbiol. 1993 Sep;9(5):923-9. doi: 10.1111/j.1365-2958.1993.tb01222.x. Mol Microbiol. 1993. PMID: 7934920 Review.
Cited by
-
Selection of biocatalysts for chemical synthesis.Proc Natl Acad Sci U S A. 2006 Feb 7;103(6):1693-8. doi: 10.1073/pnas.0504733102. Epub 2006 Jan 30. Proc Natl Acad Sci U S A. 2006. PMID: 16446453 Free PMC article.
-
Expression of the nitroarene dioxygenase genes in Comamonas sp. strain JS765 and Acidovorax sp. strain JS42 is induced by multiple aromatic compounds.J Bacteriol. 2003 Jul;185(13):3895-904. doi: 10.1128/JB.185.13.3895-3904.2003. J Bacteriol. 2003. PMID: 12813084 Free PMC article.
-
Generation of novel bacterial regulatory proteins that detect priority pollutant phenols.Appl Environ Microbiol. 2000 Jan;66(1):163-9. doi: 10.1128/AEM.66.1.163-169.2000. Appl Environ Microbiol. 2000. PMID: 10618218 Free PMC article.
-
Tracing explosives in soil with transcriptional regulators of Pseudomonas putida evolved for responding to nitrotoluenes.Microb Biotechnol. 2008 May;1(3):236-46. doi: 10.1111/j.1751-7915.2008.00027.x. Microb Biotechnol. 2008. PMID: 21261843 Free PMC article.
-
Characterisation of the putative effector interaction site of the regulatory HbpR protein from Pseudomonas azelaica by site-directed mutagenesis.PLoS One. 2011 Feb 17;6(2):e16539. doi: 10.1371/journal.pone.0016539. PLoS One. 2011. PMID: 21379585 Free PMC article.
References
-
- Abril M A, Buck M, Ramos J L. Activation of the Pseudomonas TOL plasmid upper pathway operon. Identification of binding sites for the positive regulator XylR and for integration host factor protein. J Biol Chem. 1991;266:15832–15838. - PubMed
-
- Austin S, Lambert J. Purification and in vitro activity of a truncated form of AnfA, the transcriptional activator protein of alternative nitrogenase from Azotobacter vinelandii. J Biol Chem. 1994;269:18141–18148. - PubMed
-
- Bally M, Wilberg E, Kuhni M, Egli T. Growth and regulation of enzyme synthesis in the nitrilotriacetic acid (NTA)-degrading bacterium Chelatobacter heintzii ATCC 29600. Microbiology. 1994;140:1927–1936. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

