Optimization of bacteriocin release protein (BRP)-mediated protein release by Escherichia coli: random mutagenesis of the pCloDF13-derived BRP gene to uncouple lethality and quasi-lysis from protein release
- PMID: 9464372
- PMCID: PMC106056
- DOI: 10.1128/AEM.64.2.392-398.1998
Optimization of bacteriocin release protein (BRP)-mediated protein release by Escherichia coli: random mutagenesis of the pCloDF13-derived BRP gene to uncouple lethality and quasi-lysis from protein release
Abstract
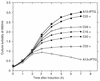
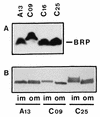
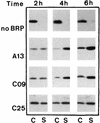
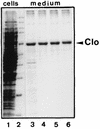
Bacteriocin release proteins (BRPs) can be used for the release of heterologous proteins from the Escherichia coli periplasm into the culture medium. However, high-level expression of BRP causes apparent lysis of the host cells in liquid cultures (quasi-lysis) and inhibition of growth on broth agar plates (lethality). To optimize BRP-mediated protein release, the pCloDF13 BRP gene was subjected to random mutagenesis by using PCR techniques. Mutated BRPs with a strongly reduced capacity to cause growth inhibition on broth agar plates were selected, analyzed by nucleotide sequencing, and further characterized by performing growth and release experiments in liquid cultures. A subset of these BRP derivatives did not cause quasi-lysis and had only a small effect on growth but still functioned in the release of the periplasmic protein beta-lactamase and the periplasmic K88 molecular chaperone FaeE and in the release of the bacteriocin cloacin DF13 into the culture medium. These BRP derivatives can be more efficiently used for extracellular production of proteins by E. coli than can the original BRP.
Figures



Similar articles
-
pCloDF13-encoded bacteriocin release proteins with shortened carboxyl-terminal segments are lipid modified and processed and function in release of cloacin DF13 and apparent host cell lysis.J Bacteriol. 1989 May;171(5):2673-9. doi: 10.1128/jb.171.5.2673-2679.1989. J Bacteriol. 1989. PMID: 2651413 Free PMC article.
-
Effect of a mutation preventing lipid modification on localization of the pCloDF13-encoded bacteriocin release protein and on release of cloacin DF13.J Bacteriol. 1988 Sep;170(9):4153-60. doi: 10.1128/jb.170.9.4153-4160.1988. J Bacteriol. 1988. PMID: 3045086 Free PMC article.
-
Uncoupling of synthesis and release of cloacin DF13 and its immunity protein by Escherichia coli.Mol Gen Genet. 1987 Jan;206(1):126-32. doi: 10.1007/BF00326547. Mol Gen Genet. 1987. PMID: 3553860
-
Functioning of a hybrid BRP-beta-lactamase protein in the release of cloacin DF13 and lysis of Escherichia coli cells.FEMS Microbiol Lett. 1989 Mar;49(1):25-31. doi: 10.1016/0378-1097(89)90336-4. FEMS Microbiol Lett. 1989. PMID: 2656396
-
Bacteriocin release proteins: mode of action, structure, and biotechnological application.FEMS Microbiol Rev. 1995 Dec;17(4):381-99. doi: 10.1111/j.1574-6976.1995.tb00221.x. FEMS Microbiol Rev. 1995. PMID: 8845188 Review.
Cited by
-
Recombinant polypeptide production in E. coli: towards a rational approach to improve the yields of functional proteins.Microb Cell Fact. 2013 Nov 1;12:101. doi: 10.1186/1475-2859-12-101. Microb Cell Fact. 2013. PMID: 24176192 Free PMC article. Review.
-
Engineering the flagellar type III secretion system: improving capacity for secretion of recombinant protein.Microb Cell Fact. 2019 Jan 18;18(1):10. doi: 10.1186/s12934-019-1058-4. Microb Cell Fact. 2019. PMID: 30657054 Free PMC article.
References
-
- Boyko W L, Ganshow R E. Rapid identification of Escherichia coli transformed by pBR322 carrying inserts at the PstI site. Anal Biochem. 1982;122:85–88. - PubMed
-
- Cavard D. Truncated mutants of the colicin A lysis protein are acylated and processed when overproduced. FEMS Microbiol Lett. 1994;117:169–174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

