Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene
- PMID: 9464384
- PMCID: PMC106072
- DOI: 10.1128/AEM.64.2.496-503.1998
Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene
Abstract
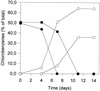
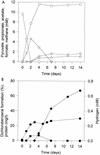
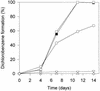
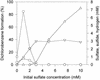
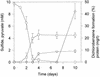
A bacterial mixed culture reductively dechlorinating trichlorobenzenes was established in a defined, synthetic mineral medium without any complex additions and with pyruvate as the carbon and energy source. The culture was maintained over 39 consecutive transfers of small inocula into fresh media, enriching the dechlorinating activity. In situ probing with fluorescence-labeled rRNA-targeted oligonucleotide probes revealed that two major subpopulations within the microbial consortium were phylogenetically affiliated with a sublineage within the Desulfovibrionaceae and the gamma subclass of Proteobacteria. The bacterial consortium grew by fermentation of pyruvate, forming acetate, propionate, CO2, formate, and hydrogen. Acetate and propionate supported neither the reduction of trichlorobenzenes nor the reduction of sulfate when sulfate was present. Hydrogen and formate were used for sulfate reduction to sulfide. Sulfate strongly inhibited the reductive dechlorination of trichlorobenzenes. However, when sulfate was depleted in the medium due to sulfate reduction, dechlorination of trichlorobenzenes started. Similar results were obtained when sulfite was present in the cultures. Molybdate at a concentration of 1 mM strongly inhibited the dechlorination of trichlorobenzenes. Cultures supplied with molybdate plus sulfate did not reduce sulfate, but dechlorination of trichlorobenzenes occurred. Supplementation of electron-depleted cultures with various electron sources demonstrated that formate was used as a direct electron donor for reductive dechlorination, whereas hydrogen was not.
Figures
References
-
- Adrian L, Manz W, Szewzyk U, Görisch H. Etablierung einer stabilen, Trichlorbenzol dechlorierenden Mischkultur und deren partielle Populationsbeschreibung mit Hilfe rRNA-gerichteter Oligonukleotidsonden. GWF Wasser-Abwasser. 1996;137:612–618.
-
- Bergmeyer H U. Methods of enzymatic analysis. 3rd ed. Weinheim, Germany: Verlag Chemie; 1984.
-
- Beurskens J E M, Dekker C G C, van den Heuvel H, Swart M, de Wolf J, Dolfing J. Dechlorination of chlorinated benzenes by an anaerobic microbial consortium that selectively mediates the thermodynamic most favorable reactions. Environ Sci Technol. 1994;28:701–706. - PubMed
-
- Bosma T N P, van der Meer J R, Schraa G, Tros M E, Zehnder A J B. Reductive dechlorination of all trichloro- and dichlorobenzene isomers. FEMS Microbiol Ecol. 1988;53:223–229.
-
- Bouchard B, Beaudet R, Villemur R, McSween G, Lépine F, Bisaillon J-G. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int J Syst Bacteriol. 1996;46:1010–1015. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

