Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp
- PMID: 9464408
- PMCID: PMC106102
- DOI: 10.1128/AEM.64.2.681-687.1998
Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp
Abstract
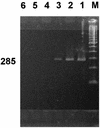
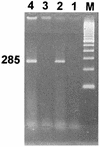
Because biochemical testing and 16S rRNA sequence analysis have proven inadequate for the differentiation of Vibrio parahaemolyticus from closely related species, we employed the gyrase B gene (gyrB) as a molecular diagnostic probe. The gyrB genes of V. parahaemolyticus and closely related Vibrio alginolyticus were cloned and sequenced. Oligonucleotide PCR primers were designed for the amplification of a 285-bp fragment from within gyrB specific for V. parahaemolyticus. These primers recognized 117 of 117 reference and wild-type V. parahaemolyticus strains, whereas amplification did not occur when 90 strains of 37 other Vibrio species or 60 strains representing 34 different nonvibrio species were tested. In 100-microliter PCR mixtures, the lower detection limits were 5 CFU for live cells and 4 pg for purified DNA. The possible application of gyrB primers for the routine identification of V. parahaemolyticus in food was examined. We developed and tested a procedure for the specific detection of the target organism in shrimp consisting of an 18-h preenrichment followed by PCR amplification of the 285-bp V. parahaemolyticus-specific fragment. This method enabled us to detect an initial inoculum of 1.5 CFU of V. parahaemolyticus cells per g of shrimp homogenate. By this approach, we were able to detect V. parahaemolyticus in all of 27 shrimp samples artificially inoculated with this bacterium. We present here a rapid, reliable, and sensitive protocol for the detection of V. parahaemolyticus in shrimp.
Figures



Similar articles
-
Cloning and nucleotide sequence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B. anthracis and their application to the detection of B. cereus in rice.Appl Environ Microbiol. 1999 Apr;65(4):1483-90. doi: 10.1128/AEM.65.4.1483-1490.1999. Appl Environ Microbiol. 1999. PMID: 10103241 Free PMC article.
-
[Quantitative detection of Vibrio parahaemolyticus by real-time TaqMan PCR].Wei Sheng Wu Xue Bao. 2005 Aug;45(4):638-42. Wei Sheng Wu Xue Bao. 2005. PMID: 16245889 Chinese.
-
Sequence of a cloned pR72H fragment and its use for detection of Vibrio parahaemolyticus in shellfish with the PCR.Appl Environ Microbiol. 1995 Apr;61(4):1311-7. doi: 10.1128/aem.61.4.1311-1317.1995. Appl Environ Microbiol. 1995. PMID: 7747952 Free PMC article.
-
Multiplex PCR assays for the detection of Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae with an internal amplification control.Diagn Microbiol Infect Dis. 2014 Jun;79(2):115-8. doi: 10.1016/j.diagmicrobio.2014.03.012. Epub 2014 Mar 18. Diagn Microbiol Infect Dis. 2014. PMID: 24731836
-
Analysis of gyrB and toxR gene sequences of Vibrio hollisae and development of gyrB- and toxR-targeted PCR methods for isolation of V. hollisae from the environment and its identification.Appl Environ Microbiol. 2000 Aug;66(8):3506-14. doi: 10.1128/AEM.66.8.3506-3514.2000. Appl Environ Microbiol. 2000. PMID: 10919814 Free PMC article.
Cited by
-
Development of a real-time resistance measurement for Vibrio parahaemolyticus detection by the lecithin-dependent hemolysin gene.PLoS One. 2013 Aug 26;8(8):e72342. doi: 10.1371/journal.pone.0072342. eCollection 2013. PLoS One. 2013. PMID: 23991096 Free PMC article.
-
Development of a rapid PCR protocol to detect Vibrio parahaemolyticus in clams.J Food Sci Technol. 2018 Feb;55(2):749-759. doi: 10.1007/s13197-017-2986-9. Epub 2017 Dec 22. J Food Sci Technol. 2018. PMID: 29391640 Free PMC article.
-
Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene.J Clin Microbiol. 1999 Apr;37(4):1173-7. doi: 10.1128/JCM.37.4.1173-1177.1999. J Clin Microbiol. 1999. PMID: 10074546 Free PMC article.
-
Quantitative real-time PCR and fluorescence in situ hybridization approaches for enumerating Brevundimonas diminuta in drinking water.J Ind Microbiol Biotechnol. 2010 Sep;37(9):909-18. doi: 10.1007/s10295-010-0738-1. Epub 2010 May 22. J Ind Microbiol Biotechnol. 2010. PMID: 20495940
-
The heat shock protein 70 gene as a new alternative molecular marker for the taxonomic identification of Streptomyces strains.AMB Express. 2018 Sep 10;8(1):144. doi: 10.1186/s13568-018-0674-4. AMB Express. 2018. PMID: 30203150 Free PMC article.
References
-
- Baba K, Shirai H, Terai A, Takeda Y, Nishibuchi M. Analysis of the tdh gene cloned from a tdh gene- and trh gene-positive strain of Vibrio parahaemolyticus. Microbiol Immunol. 1991;35:253–258. - PubMed
-
- Baumann P, Schubert R H W. Family II. Vibrionaceae Veron 1965, 5245. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 516–550.
-
- Honda T, Iida T. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct hemolysin and related hemolysins. Rev Med Microbiol. 1993;4:106–113.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

