A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics
- PMID: 9465037
- PMCID: PMC19045
- DOI: 10.1073/pnas.95.4.1460
A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics
Abstract
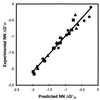
A unified view of polymer, dumbbell, and oligonucleotide nearest-neighbor (NN) thermodynamics is presented. DNA NN DeltaG degrees 37 parameters from seven laboratories are presented in the same format so that careful comparisons can be made. The seven studies used data from natural polymers, synthetic polymers, oligonucleotide dumbbells, and oligonucleotide duplexes to derive NN parameters; used different methods of data analysis; used different salt concentrations; and presented the NN thermodynamics in different formats. As a result of these differences, there has been much confusion regarding the NN thermodynamics of DNA polymers and oligomers. Herein I show that six of the studies are actually in remarkable agreement with one another and explanations are provided in cases where discrepancies remain. Further, a single set of parameters, derived from 108 oligonucleotide duplexes, adequately describes polymer and oligomer thermodynamics. Empirical salt dependencies are also derived for oligonucleotides and polymers.
Figures

References
-
- Crothers D M, Zimm B H. J Mol Biol. 1964;9:1–9. - PubMed
-
- DeVoe H, Tinoco I., Jr J Mol Biol. 1962;4:500–517. - PubMed
-
- Gray D M, Tinoco I., Jr Biopolymers. 1970;9:223–244.
-
- Borer P N, Dengler B, Tinoco I, Jr, Uhlenbeck O C. J Mol Biol. 1974;86:843–853. - PubMed
-
- Tinoco I, Jr, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Nat New Biol. 1973;246:40–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

