Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation
- PMID: 9465073
- PMCID: PMC19140
- DOI: 10.1073/pnas.95.4.1663
Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation
Abstract
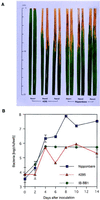
The Xa1 gene in rice confers resistance to Japanese race 1 of Xanthomonas oryzae pv. oryzae, the causal pathogen of bacterial blight (BB). We isolated the Xa1 gene by a map-based cloning strategy. The deduced amino acid sequence of the Xa1 gene product contains nucleotide binding sites (NBS) and a new type of leucine-rich repeats (LRR); thus, Xa1 is a member of the NBS-LRR class of plant disease-resistance genes, but quite different from Xa21, another BB-resistance gene isolated from rice. Interestingly, Xa1 gene expression was induced on inoculation with a bacterial pathogen and wound, unlike other isolated resistance genes in plants, which show constitutive expression. The induced expression may be involved in enhancement of resistance against the pathogen.
Figures


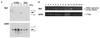
References
-
- Mew T W. Annu Rev Phytopathol. 1987;25:359–382.
-
- Kinoshita T. Rice Genet Newsl. 1992;8:2–37.
-
- Khush G S, Mackill D J, Sidhu G S. Bacterial Blight of Rice. Manila, Philippines: International Rice Research Institute; 1989. pp. 207–217.
-
- Sakaguchi S. Bull Natl Inst Agr Sci Ser. 1967;D16:1–18. (in Japanese with English summary).
-
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

