Negative regulation of granulocytic differentiation in the myeloid precursor cell line 32Dcl3 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1/ETO
- PMID: 9465099
- PMCID: PMC19195
- DOI: 10.1073/pnas.95.4.1812
Negative regulation of granulocytic differentiation in the myeloid precursor cell line 32Dcl3 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1/ETO
Abstract
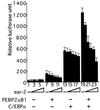
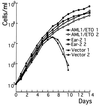
The polyomavirus enhancer binding protein 2alphaB (AML1/PEBP2alphaB/Cbfa2) plays a pivotal role in granulocyte colony-stimulating factor (G-CSF)-mediated differentiation of a myeloid progenitor cell line, 32Dc13. In this article, we report the identification of a PEBP2alphaB interacting protein, Ear-2, an orphan member of the nuclear hormone receptor superfamily that directly binds to and can inhibit the function of PEBP2alphaB. Ear-2 is expressed in proliferating 32Dc13 cells in presence of interleukin 3 but is down-regulated during differentiation induced by G-CSF. Interestingly, AML1/ETO(MTG8), a leukemogenic chimeric protein can block the differentiation of 32Dc13 cells, which is accompanied by the sustained expression of ear-2. Overexpression of Ear-2 can prevent G-CSF-induced differentiation, strongly suggesting that ear-2 is a key negative regulator of granulocytic differentiation. Our results indicate that a dynamic balance existing between PEBP2alphaB and Ear-2 appears to determine the choice between growth or differentiation for myeloid cells.
Figures


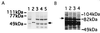


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

