Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis
- PMID: 9466739
- PMCID: PMC104540
- DOI: 10.1128/JCM.36.2.345-351.1998
Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis
Abstract
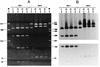
Urogenital isolates (n = 93) of Chlamydia trachomatis were differentiated into serovars and variants by serotyping with monoclonal antibodies and genotyping by restriction fragment length polymorphism (RFLP) analysis of the PCR-amplified omp1 gene, respectively. The types of 87 of the 93 isolates (94%) were identical, as determined by both methods. Among these 87 isolates, 3 isolates were identified as the recently described new serovariant Ga/IOL-238 by omp1 nucleotide sequence analysis of the variable domains. Of the remaining six isolates, three isolates serotyped as both L2 and Ba but were identified as Ba/A-7 by genotyping by RFLP analysis of omp1. The omp1 nucleotide sequences of variable domains VD1, VD2, and VD4 of these urogenital Ba strains were identical to the sequences of the variable domains of Ba/J160, an ocular Ba type. The three remaining isolates were serotyped as J, but the patterns obtained by RFLP analysis of omp1, which were identical for the three isolates, differed from that of prototype serovar J/UW36. omp1 nucleotide sequence analysis revealed that these strains are genovariants of serovar J/UW36. Nucleotide sequence differences between serovar J/UW36 and this J genovariant, designated Jv, were found in both variable and constant domains. In conclusion, this study shows that the PCR-based genotyping of clinical C. trachomatis isolates by RFLP analysis of omp1 has a higher discriminatory power and is more convenient than serotyping. Variants of C. trachomatis serovars Ba, G, and J were identified and characterized.
Figures

References
-
- Dean D, Schachter J, Dawson C R, Stephens S. Comparison of the major outer membrane protein variant sequence regions of B/Ba isolates: a molecular epidemiologic approach to Chlamydia trachomatis infections. J Infect Dis. 1992;166:383–392. - PubMed
-
- Dean D. Molecular characterization of a new Chlamydia trachomatis serological variant from a trachoma endemic region of Africa. In: Orfila J, Byrne G I, Chernesky M A, et al., editors. Chlamydial infections. Proceedings of the Eighth International Symposium on Human Chlamydial Infections, Chantilly, France. Bologna, Italy: Società Editrice Esculapio; 1994. pp. 259–262.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

