Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy
- PMID: 9466740
- PMCID: PMC104541
- DOI: 10.1128/JCM.36.2.352-357.1998
Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy
Abstract
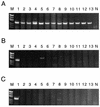
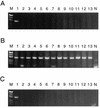
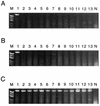
We have developed a method for determining the serotypes of poliovirus isolates by PCR. Three sets of serotype-specific antisense PCR-initiating primers (primers seroPV1A, seroPV2A, and seroPV3A) were designed to pair with codons of VP1 amino acid sequences that are conserved within but that differ across serotypes. The sense polarity primers (primers seroPV1S, seroPV2S, and seroPV3S) matched codons of more conserved capsid sequences. The primers contain mixed-base and deoxyinosine residues to compensate for the high rate of degeneracy of the targeted codons. The serotypes of all polioviruses tested (48 vaccine-related isolates and 110 diverse wild isolates) were correctly identified by PCR with the serotype-specific primers. None of the genomic sequences of 49 nonpolio enterovirus reference strains were amplified under equivalent reaction conditions with any of the three primer sets. These primers are useful for the rapid screening of poliovirus isolates and for determining the compositions of cultures containing mixtures of poliovirus serotypes.
Figures


References
-
- Balanant J, Guillot S, Candréa A, Delpeyroux F, Crainic R. The natural genome variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology. 1991;184:645–654. - PubMed
-
- Birmingham, M. E., R. W. Linkins, B. P. Hull, and H. F. Hull. 1997. Poliomyelitis surveillance: the compass for eradication. J. Infect. Dis. 175(Suppl. 1):S146–S150. - PubMed
-
- Bodian D, Morgan I M, Howe H A. Differentiation of types of poliomyelitis viruses. Am J Hyg. 1949;49:234–245. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

