Detection of Tritrichomonas foetus by PCR and DNA enzyme immunoassay based on rRNA gene unit sequences
- PMID: 9466768
- PMCID: PMC104569
- DOI: 10.1128/JCM.36.2.513-519.1998
Detection of Tritrichomonas foetus by PCR and DNA enzyme immunoassay based on rRNA gene unit sequences
Abstract
Tritrichomonas foetus is the causative agent of bovine tritrichomonosis, a sexually transmitted disease leading to infertility and abortion. Diagnosis is hampered by putative contamination of samples with intestinal or coprophilic trichomonadid protozoa which might be mistaken for T. foetus. Therefore, we developed a PCR test optimized for applicability in routine diagnosis. Amplification is based upon primers TFR3 and TFR4 directed to the rRNA gene units of T. foetus. In order to avoid potential carryover contamination by products of previous amplification reactions, conditions were adapted to the use of the uracil DNA glycosylase system. Furthermore, documentation and interpretation of results were facilitated by including a DNA enzyme immunoassay for the detection of amplification products. Specificity was confirmed with genomic material from different related trichomonadid protozoa. The high sensitivity of the test allowed the detection of a single T. foetus organism in diagnostic culture medium or about 50 parasites per ml of preputial washing fluid. The present methods are thus proposed as (i) confirmatory tests for microscopic diagnosis following diagnostic in vitro cultivation and (ii) a direct T. foetus screening test with diagnostic samples.
Figures


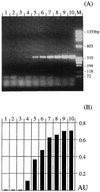

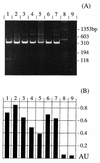
Similar articles
-
PCR detection of Tritrichomonas foetus in preputial bull fluid without prior DNA isolation.Vet Parasitol. 2006 Mar 31;136(3-4):357-61. doi: 10.1016/j.vetpar.2005.11.016. Epub 2005 Dec 28. Vet Parasitol. 2006. PMID: 16386373
-
Single-tube nested PCR for detection of tritrichomonas foetus in feline feces.J Clin Microbiol. 2002 Nov;40(11):4126-30. doi: 10.1128/JCM.40.11.4126-4130.2002. J Clin Microbiol. 2002. PMID: 12409385 Free PMC article.
-
Intestinal Tritrichomonas foetus infection in cats in Switzerland detected by in vitro cultivation and PCR.Parasitol Res. 2009 Mar;104(4):783-8. doi: 10.1007/s00436-008-1255-2. Epub 2008 Nov 8. Parasitol Res. 2009. PMID: 18998166
-
Host-parasite interaction in bovine infection with Tritrichomonas foetus.Microbes Infect. 1999 Aug;1(10):807-16. doi: 10.1016/s1286-4579(99)80083-5. Microbes Infect. 1999. PMID: 10816086 Review.
-
What is known about Tritrichomonas foetus infection in cats?Rev Bras Parasitol Vet. 2019 Jan-Mar;28(1):1-11. doi: 10.1590/S1984-29612019005. Epub 2019 Mar 11. Rev Bras Parasitol Vet. 2019. PMID: 30892464 Review.
Cited by
-
Prevalence of Tritrichomonas foetus Among Cats in Poland Between 2020 and 2024.Pathogens. 2025 May 7;14(5):458. doi: 10.3390/pathogens14050458. Pathogens. 2025. PMID: 40430778 Free PMC article.
-
Systematic Review of Vaccine Strategies Against Tritrichomonas foetus Infection in Cattle: Insights, Challenges, and Prospects.Parasite Immunol. 2025 Jan;47(1):e70003. doi: 10.1111/pim.70003. Parasite Immunol. 2025. PMID: 39838701 Free PMC article.
-
Neurotoxicosis in 4 cats receiving ronidazole.J Vet Intern Med. 2007 Mar-Apr;21(2):328-31. doi: 10.1892/0891-6640(2007)21[328:nicrr]2.0.co;2. J Vet Intern Med. 2007. PMID: 17427396 Free PMC article. No abstract available.
-
Diagnosis of trichomoniasis in 'virgin' bulls by culture and polymerase chain reaction.Can Vet J. 2003 Sep;44(9):732-4. Can Vet J. 2003. PMID: 14524627 Free PMC article.
-
An improved polymerase chain reaction assay for the detection of Tritrichomonas foetus in cattle.Can Vet J. 2002 Mar;43(3):213-6. Can Vet J. 2002. PMID: 11901595 Free PMC article.
References
-
- Appell L H, Mickelsen W D, Thomas M W, Harmon W M. A comparison of techniques used for the diagnosis of Tritrichomonas foetus infections in beef bulls. Agri Pract. 1993;14:30–34.
-
- BonDurant R H, Anderson M L, Blanchard P, Hird D, Danaye-Elmi C, Palmer C, Sischo W M, Suther D, Utterback W, Weigler B J. Prevalence of trichomoniasis among California beef herds. J Am Vet Med Assoc. 1990;196:1590–1593. - PubMed
-
- Borchardt K A, Norman B B, Thomas M W, Harmon W M. Evaluation of a new culture method for diagnosing Tritrichomonas foetus infection. Vet Med. 1992;1992:104–112.
-
- Chakrabarti D, Dame J B, Gutell R R, Yowell C A. Characterization of the rDNA unit and sequence analysis of the small subunit rRNA and 5.8S rRNA genes from Tritrichomonas foetus. Mol Biochem Parasitol. 1992;52:75–84. - PubMed
-
- Corbeil L B. Vaccination strategies against Tritrichomonas foetus. Parasitol Today. 1994;10:103–106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

