Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157
- PMID: 9466788
- PMCID: PMC104589
- DOI: 10.1128/JCM.36.2.598-602.1998
Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157
Abstract
Shiga toxigenic Escherichia coli (STEC) comprises a diverse group of organisms capable of causing severe gastrointestinal disease in humans. Within the STEC family, certain strains appear to be of greater virulence for humans, for example, those belonging to serogroups O111 and O157 and those with particular combinations of other putative virulence traits. We have developed two multiplex PCR assays for the detection and genetic characterization of STEC in cultures of feces or foodstuffs. Assay 1 utilizes four PCR primer pairs and detects the presence of stx1, stx2 (including variants of stx2), eaeA, and enterohemorrhagic E. coli hlyA, generating amplification products of 180, 255, 384, and 534 bp, respectively. Assay 2 uses two primer pairs specific for portions of the rfb (O-antigen-encoding) regions of E. coli serotypes O157 and O111, generating PCR products of 259 and 406 bp, respectively. The two assays were validated by testing 52 previously characterized STEC strains and observing 100% agreement with previous results. Moreover, assay 2 did not give a false-positive O157 reaction with enteropathogenic E. coli strains belonging to clonally related serogroup O55. Assays 1 and 2 detected STEC of the appropriate genotype in primary fecal cultures from five patients with hemolytic-uremic syndrome and three with bloody diarrhea. Thirty-one other primary fecal cultures from patients without evidence of STEC infection were negative.
Figures
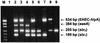



Similar articles
-
Semi-automated fluorogenic PCR assays (TaqMan) forrapid detection of Escherichia coli O157:H7 and other shiga toxigenic E. coli.Mol Cell Probes. 1999 Aug;13(4):291-302. doi: 10.1006/mcpr.1999.0251. Mol Cell Probes. 1999. PMID: 10441202
-
Direct detection of Shiga toxigenic Escherichia coli strains belonging to serogroups O111, O157, and O113 by multiplex PCR.J Clin Microbiol. 1999 Oct;37(10):3362-5. doi: 10.1128/JCM.37.10.3362-3365.1999. J Clin Microbiol. 1999. PMID: 10488207 Free PMC article.
-
Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction.J Food Prot. 2002 Sep;65(9):1371-80. doi: 10.4315/0362-028x-65.9.1371. J Food Prot. 2002. PMID: 12233845
-
Detection of shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR.Appl Environ Microbiol. 1999 Feb;65(2):868-72. doi: 10.1128/AEM.65.2.868-872.1999. Appl Environ Microbiol. 1999. PMID: 9925634 Free PMC article.
-
Multiplex real-time PCR assay for detection of Escherichia coli O157:H7 and screening for non-O157 Shiga toxin-producing E. coli.BMC Microbiol. 2017 Nov 9;17(1):215. doi: 10.1186/s12866-017-1123-2. BMC Microbiol. 2017. PMID: 29121863 Free PMC article.
Cited by
-
Incidence and virulence determinants of verocytotoxin-producing Escherichia coli infections in the Brussels-Capital Region, Belgium, in 2008-2010.J Clin Microbiol. 2012 Apr;50(4):1336-45. doi: 10.1128/JCM.05317-11. Epub 2012 Jan 11. J Clin Microbiol. 2012. PMID: 22238434 Free PMC article.
-
Characterization of novel bacteriophage phiC119 capable of lysing multidrug-resistant Shiga toxin-producing Escherichia coli O157:H7.PeerJ. 2016 Sep 13;4:e2423. doi: 10.7717/peerj.2423. eCollection 2016. PeerJ. 2016. PMID: 27672499 Free PMC article.
-
Extended-spectrum-β-lactamase-producing Escherichia coli isolate possessing the Shiga toxin gene (stx1) belonging to the O64 serogroup associated with human disease in India.J Clin Microbiol. 2013 Jun;51(6):2008-9. doi: 10.1128/JCM.00575-13. Epub 2013 Apr 10. J Clin Microbiol. 2013. PMID: 23576543 Free PMC article. No abstract available.
-
Whole-Genome Characterization and Strain Comparison of VT2f-Producing Escherichia coli Causing Hemolytic Uremic Syndrome.Emerg Infect Dis. 2016 Dec;22(12):2078-2086. doi: 10.3201/eid2212.160017. Epub 2016 Dec 15. Emerg Infect Dis. 2016. PMID: 27584691 Free PMC article.
-
Enteric bacterial pathogens in children with diarrhea in Niger: diversity and antimicrobial resistance.PLoS One. 2015 Mar 23;10(3):e0120275. doi: 10.1371/journal.pone.0120275. eCollection 2015. PLoS One. 2015. PMID: 25799400 Free PMC article.
References
-
- Barrett T J, Kaper J B, Jerse A E, Wachsmuth I K. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J Infect Dis. 1992;165:979–980. - PubMed
-
- Bastin D A, Reeves P R. Sequence analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene. 1995;164:17–23. - PubMed
-
- Begum D, Jackson M P. Direct detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Mol Cell Probes. 1995;9:259–264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

