High mobility group protein-1 (HMG-1) is a unique activator of p53
- PMID: 9472015
- PMCID: PMC316524
- DOI: 10.1101/gad.12.4.462
High mobility group protein-1 (HMG-1) is a unique activator of p53
Abstract
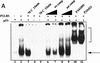
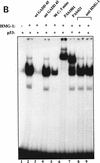
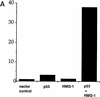
The binding of p53 protein to DNA is stimulated by its interaction with covalent as well as noncovalent modifiers. We report the identification of a factor from HeLa nuclear extracts that activates p53 DNA binding. This factor was purified to homogeneity and identified as the high mobility group protein, HMG-1. HMG-1 belongs to a family of highly conserved chromatin-associated nucleoproteins that bend DNA and facilitate the binding of various transcription factors to their cognate DNA sequences. We demonstrate that recombinant His-tagged HMG-1 enhances p53 DNA binding in vitro and also that HMG-1 and p53 can interact directly in vitro. Unexpectedly, HMG-1 also stimulates DNA binding by p53Delta30, a carboxy-terminally deleted form of the protein that is considered to be constitutively active, suggesting that HMG-1 stimulates p53 by a mechanism that is distinct from other known activators of p53. Finally, using transient transfection assays we show that HMG-1 can increase p53 and p53Delta30-mediated transactivation in vivo. HMG-1 promotes the assembly of higher order p53 nucleoprotein structures, and these data, along with the fact that HMG-1 is capable of bending DNA, suggest that HMG-1 may activate p53 DNA binding by a novel mechanism involving a structural change in the target DNA.
Figures








References
-
- Agarwal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. - PubMed
-
- Bazett-Jones DP, LeBlanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
