tRNA(Arg) (fimU) and expression of SEF14 and SEF21 in Salmonella enteritidis
- PMID: 9473037
- PMCID: PMC106962
- DOI: 10.1128/JB.180.4.840-845.1998
tRNA(Arg) (fimU) and expression of SEF14 and SEF21 in Salmonella enteritidis
Abstract
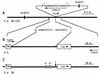
A Tn10 insertion affecting SEF14 fimbrial synthesis in Salmonella enteritidis was located 13 bp upstream of a gene designated fimU. The 77-bp DNA sequence of fimU from S. enteritidis was identical to that of fimU encoding tRNA(Arg) (UCU) from Salmonella typhimurium and 96% identical to that of the Escherichia coli argU homolog. Furthermore, the open reading frame adjacent to and overlapping the 3' end of fimU was similar to the prophage DLP12 integrase gene. The fimU-encoded transcript comigrated with total cellular tRNA and was predicted to form a tRNA-like cloverleaf structure containing the arginine anticodon UCU. Thus, fimU encoded a tRNA(Arg) specific for the rare codon AGA. fimU mapped to the SEF21 fim operon located 15 C's from the sef14 gene cluster. Although fimU was located within the SEF21 fim gene cluster, the fimU Tn10 insertion mutant of S. enteritidis was found to be defective in SEF14 as well as SEF21 (type 1) fimbria production. SEF17 and SEF18 fimbria production was not affected. Complementation of this mutant with plasmid-borne fimU restored normal production of the fimbrins SefA and FimA as well as their respective fimbriae SEF14 and SEF21. This is the first description of tRNA simultaneously controlling the production of two distinct fimbriae.
Figures


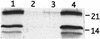


References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Chen G-F, Inouye M. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev. 1994;8:2641–2652. - PubMed
-
- Clouthier S C. Ph.D. thesis. Victoria, Canada: University of Victoria; 1995.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

