Cleaving DNA with DNA
- PMID: 9482868
- PMCID: PMC19303
- DOI: 10.1073/pnas.95.5.2233
Cleaving DNA with DNA
Abstract
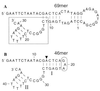
A DNA structure is described that can cleave single-stranded DNA oligonucleotides in the presence of ionic copper. This "deoxyribozyme" can self-cleave or can operate as a bimolecular complex that simultaneously makes use of duplex and triplex interactions to bind and cleave separate DNA substrates. Bimolecular deoxyribozyme-mediated strand scission proceeds with a kobs of 0.2 min-1, whereas the corresponding uncatalyzed reaction could not be detected. The duplex and triplex recognition domains can be altered, making possible the targeted cleavage of single-stranded DNAs with different nucleotide sequences. Several small synthetic DNAs were made to function as simple "restriction enzymes" for the site-specific cleavage of single-stranded DNA.
Figures
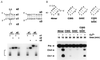
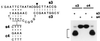


Similar articles
-
Characterization of a DNA-cleaving deoxyribozyme.Bioorg Med Chem. 2001 Oct;9(10):2589-600. doi: 10.1016/s0968-0896(01)00035-9. Bioorg Med Chem. 2001. PMID: 11557347
-
Development of a short Ca2+-dependent deoxyribozyme with RNA cleavage activity.Nucleic Acids Symp Ser. 1999;(42):281-2. doi: 10.1093/nass/42.1.281. Nucleic Acids Symp Ser. 1999. PMID: 10780489
-
Homologous recognition and triplex formation promoted by RecA protein between duplex oligonucleotides and single-stranded DNA.J Mol Biol. 1993 Jan 20;229(2):328-43. doi: 10.1006/jmbi.1993.1038. J Mol Biol. 1993. PMID: 8381491
-
Sequence-selective recognition and cleavage of double-helical DNA.Curr Opin Biotechnol. 1993 Feb;4(1):29-36. doi: 10.1016/0958-1669(93)90028-u. Curr Opin Biotechnol. 1993. PMID: 7763389 Review.
-
Single-molecule manipulation quantification of site-specific DNA binding.Curr Opin Chem Biol. 2019 Dec;53:106-117. doi: 10.1016/j.cbpa.2019.08.006. Epub 2019 Oct 31. Curr Opin Chem Biol. 2019. PMID: 31677535 Review.
Cited by
-
Construction and Application of DNAzyme-based Nanodevices.Chem Res Chin Univ. 2023;39(1):42-60. doi: 10.1007/s40242-023-2334-8. Epub 2023 Jan 16. Chem Res Chin Univ. 2023. PMID: 36687211 Free PMC article. Review.
-
Detection of intracellular microRNA-21 for cancer diagnosis by a nanosystem containing a ZnO@polydopamine and DNAzyme probe.RSC Adv. 2024 Apr 26;14(19):13351-13360. doi: 10.1039/d4ra00636d. eCollection 2024 Apr 22. RSC Adv. 2024. PMID: 38680416 Free PMC article.
-
Selective tumor cell death induced by irradiated riboflavin through recognizing DNA G-T mismatch.Nucleic Acids Res. 2017 Sep 6;45(15):8676-8683. doi: 10.1093/nar/gkx602. Nucleic Acids Res. 2017. PMID: 28911109 Free PMC article.
-
Functional nucleic acid sensors.Chem Rev. 2009 May;109(5):1948-98. doi: 10.1021/cr030183i. Chem Rev. 2009. PMID: 19301873 Free PMC article. Review. No abstract available.
-
Tethered imidazole mediated duplex stabilization and its potential for aptamer stabilization.Nucleic Acids Res. 2018 Dec 14;46(22):11671-11686. doi: 10.1093/nar/gky1062. Nucleic Acids Res. 2018. PMID: 30418582 Free PMC article.
References
-
- Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. - PubMed
-
- Gold L, Poliski B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. - PubMed
-
- Osborne S E, Ellington A D. Chem Rev. 1997;97:349–370. - PubMed
-
- Breaker R R. Curr Opin Chem Biol. 1997;1:26–31. - PubMed
-
- Burgstaller P, Famulok M. Angew Chem Int Ed Engl. 1995;34:1189–1192.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

