beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres
- PMID: 9482891
- PMCID: PMC19348
- DOI: 10.1073/pnas.95.5.2366
beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres
Abstract
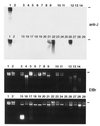
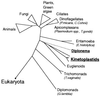
The unusual DNA base beta-D-glucosyl-hydroxymethyluracil, called "J, " replaces approximately 0.5-1% of Thy in DNA of African trypanosomes but has not been found in other organisms thus far. In Trypanosoma brucei, J is located predominantly in repetitive DNA, and its presence correlates with the silencing of telomeric genes. Using antibodies specific for J, we have developed sensitive assays to screen for J in a range of organisms and have found that J is not limited to trypanosomes that undergo antigenic variation but is conserved among Kinetoplastida. In all kinetoplastids tested, including the human pathogens Leishmania donovani and Trypanosoma cruzi, J was found to be abundantly present in the (GGGTTA)n telomere repeats. Outside Kinetoplastida, J was found only in Diplonema, a small phagotrophic marine flagellate, in which we also identified 5-MeCyt. Fractionation of Diplonema DNA showed that the two modifications are present in a common genome compartment, which suggests that they may have a similar function. Dinoflagellates appear to contain small amounts of modified bases that may be analogs of J. The evolutionary conservation of J in kinetoplastid protozoans suggests that it has a general function, repression of transcription or recombination, or a combination of both. T. brucei may have recruited J for the control of genes involved in antigenic variation.
Figures


Comment in
-
A base called J.Proc Natl Acad Sci U S A. 1998 Mar 3;95(5):2037-8. doi: 10.1073/pnas.95.5.2037. Proc Natl Acad Sci U S A. 1998. PMID: 9482833 Free PMC article. Review. No abstract available.
References
-
- Gommers-Ampt J H, van Leeuwen F, de Beer A L, Vliegenthart J F, Dizdaroglu M, Kowalak J A, Crain P F, Borst P. Cell. 1993;75:1129–1136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

