Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway
- PMID: 9488381
- PMCID: PMC108001
- DOI: 10.1128/IAI.66.3.950-958.1998
Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway
Abstract
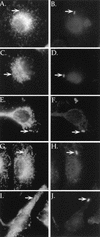
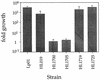
Legionella pneumophila replicates within a specialized phagosome in cultured cells, a function necessary for its pathogenicity. The replicative phagosome lacks membrane marker proteins, such as the glycoprotein LAMP-1, that are indicators of the normal endocytic pathway. We describe the isolation of several Legionella genes essential for intracellular growth and evasion of the endocytic pathway, using a genetic and cell biological approach. We screened 4,960 ethyl methanesulfonate-mutagenized colonies for defects in intracellular growth and trafficking to the replicative phagosome. Six mutant strains of L. pneumophila that had severe intracellular growth defects in mouse bone marrow-derived macrophages were identified. All six mutants were found in phagosomes that colocalized with LAMP-1, indicating defects in intracellular trafficking. The growth defects of two of these strains were complemented by molecular clones from a bank constructed from a wild-type L. pneumophila strain. The inserts from these clones are located in a region of the chromosome contiguous with several other genes essential for intracellular growth. Three mutants could be complemented by single open reading frames placed in trans, one mutant by a gene termed dotH and two additional mutants by a gene termed dotO. A deletion mutation was created in a third gene, dotI, which is located directly upstream of dotH. The delta dotI strain was also defective for intracellular growth in macrophages, and this defect was complemented by a single open reading frame in trans. Based on sequence analysis and structural predictions, possible roles of dotH, dotI, and dotO in intracellular growth are discussed.
Figures






References
-
- Andrews, H. L., J. P. Vogel, and R. R. Isberg. Unpublished data.
-
- Berger K H, Isberg R R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. - PubMed
-
- Berger K H, Merriam J J, Isberg R R. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol. 1994;14:809–822. - PubMed
-
- Brand B C, Sadosky A B, Shuman H A. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol. 1994;14:797–808. - PubMed
-
- Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–378.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

